[English] 日本語
 Yorodumi
Yorodumi- EMDB-7859: Full Length HIV-1 Envelope Glycoprotein clone PC64M18C043 in comp... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7859 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Full Length HIV-1 Envelope Glycoprotein clone PC64M18C043 in complex with PGT151 Fab and PCT64-35S Fab | |||||||||
 Map data Map data | Full Length HIV-1 Envelope Glycoprotein clone PC64M18C043 in complex with PGT151 Fab and PCT64-35S Fab, primary map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
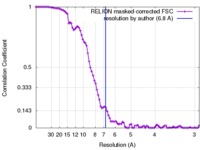
| Method | single particle reconstruction / cryo EM / Resolution: 6.8 Å | |||||||||
 Authors Authors | Berndsen ZT / Rantalainen K / Ward AB | |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2018 Journal: Cell Rep / Year: 2018Title: Co-evolution of HIV Envelope and Apex-Targeting Neutralizing Antibody Lineage Provides Benchmarks for Vaccine Design. Authors: Kimmo Rantalainen / Zachary T Berndsen / Sasha Murrell / Liwei Cao / Oluwarotimi Omorodion / Jonathan L Torres / Mengyu Wu / Jeffrey Umotoy / Jeffrey Copps / Pascal Poignard / Elise Landais ...Authors: Kimmo Rantalainen / Zachary T Berndsen / Sasha Murrell / Liwei Cao / Oluwarotimi Omorodion / Jonathan L Torres / Mengyu Wu / Jeffrey Umotoy / Jeffrey Copps / Pascal Poignard / Elise Landais / James C Paulson / Ian A Wilson / Andrew B Ward /  Abstract: Broadly neutralizing antibodies (bnAbs) targeting the HIV envelope glycoprotein (Env) typically take years to develop. Longitudinal analyses of both neutralizing antibody lineages and viruses at ...Broadly neutralizing antibodies (bnAbs) targeting the HIV envelope glycoprotein (Env) typically take years to develop. Longitudinal analyses of both neutralizing antibody lineages and viruses at serial time points during infection provide a basis for understanding the co-evolutionary contest between HIV and the humoral immune system. Here, we describe the structural characterization of an apex-targeting antibody lineage and autologous clade A viral Env from a donor in the Protocol C cohort. Comparison of Ab-Env complexes at early and late time points reveals that, within the antibody lineage, the CDRH3 loop rigidifies, the bnAb angle of approach steepens, and surface charges are mutated to accommodate glycan changes. Additionally, we observed differences in site-specific glycosylation between soluble and full-length Env constructs, which may be important for tuning optimal immunogenicity in soluble Env trimers. These studies therefore provide important guideposts for design of immunogens that prime and mature nAb responses to the Env V2-apex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7859.map.gz emd_7859.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7859-v30.xml emd-7859-v30.xml emd-7859.xml emd-7859.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7859_fsc.xml emd_7859_fsc.xml | 9 KB | Display |  FSC data file FSC data file |
| Images |  emd_7859.png emd_7859.png | 47.9 KB | ||
| Others |  emd_7859_half_map_1.map.gz emd_7859_half_map_1.map.gz emd_7859_half_map_2.map.gz emd_7859_half_map_2.map.gz | 49.5 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7859 http://ftp.pdbj.org/pub/emdb/structures/EMD-7859 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7859 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7859 | HTTPS FTP |
-Related structure data
| Related structure data |  7858C  7860C  7861C  7862C  7863C  7864C  7865C  7866C  6ca6C  6ca7C  6ca9C  6dcqC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7859.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7859.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full Length HIV-1 Envelope Glycoprotein clone PC64M18C043 in complex with PGT151 Fab and PCT64-35S Fab, primary map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.46 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
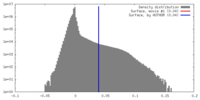
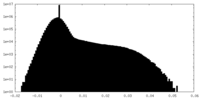
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Full Length HIV-1 Envelope Glycoprotein clone PC64M18C043 in...
| File | emd_7859_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full Length HIV-1 Envelope Glycoprotein clone PC64M18C043 in complex with PGT151 Fab and PCT64-35S Fab, half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
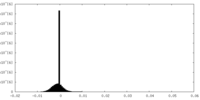
| Density Histograms |
-Half map: Full Length HIV-1 Envelope Glycoprotein clone PC64M18C043 in...
| File | emd_7859_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full Length HIV-1 Envelope Glycoprotein clone PC64M18C043 in complex with PGT151 Fab and PCT64-35S Fab, half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
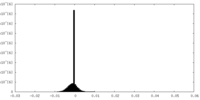
| Density Histograms |
- Sample components
Sample components
-Entire : Full Length HIV-1 Envelope Glycoprotein clone PC64M18C043 in comp...
| Entire | Name: Full Length HIV-1 Envelope Glycoprotein clone PC64M18C043 in complex with PCT64-35S Fab and PGT151 Fab |
|---|---|
| Components |
|
-Supramolecule #1: Full Length HIV-1 Envelope Glycoprotein clone PC64M18C043 in comp...
| Supramolecule | Name: Full Length HIV-1 Envelope Glycoprotein clone PC64M18C043 in complex with PCT64-35S Fab and PGT151 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293F Homo sapiens (human) / Recombinant cell: HEK293F |
-Macromolecule #1: Full Length HIV-1 Envelope Glycoprotein clone PC64M18C043
| Macromolecule | Name: Full Length HIV-1 Envelope Glycoprotein clone PC64M18C043 type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKVKGIQMNC QHLLRWGTMI LGLIIICSAA NNLWVTVYYG VPVWRDAETT LFCASDAKAY DTEVHNVWAT HACVPTDPSP QEIHLANVTE KFDMWKNSMV EQMHTDIISL WDESLKPCVK LTPLCITLNC TNITRNVTGG NLTEEGKEEL KNCSFNATTE LRDKIQKVHS ...String: MKVKGIQMNC QHLLRWGTMI LGLIIICSAA NNLWVTVYYG VPVWRDAETT LFCASDAKAY DTEVHNVWAT HACVPTDPSP QEIHLANVTE KFDMWKNSMV EQMHTDIISL WDESLKPCVK LTPLCITLNC TNITRNVTGG NLTEEGKEEL KNCSFNATTE LRDKIQKVHS LFYRLDLVEL NEGNSSDSNT SMYRLINCNT SAITQACPKV SFEPIPIHYC APAGFAILKC REKEFNGTGP CKKVSTVQCT HGIKPVVSTQ LLLNGSLAEG KVKIRCENIS NNAKTILVQL TTPVRINCTR PSNNTRTSIR IGPGQSFYAT GDIIGDIRKA YCNVSESEWK EALGKVVEQL RNHFNKTITF ASSSGGDLEI TTHSFNCGGE FFYCNTSSLF NSTWDGNSAT NSTQVPNGTI TLPCRIKQII NMWQRTGQAM YAPPIPGKIR CDSNITGLIL IRDGGNNNNE SETFRPGGGD MRNNWRSELY KYKVVKIDPL GVAPTGAKRR VVEREKRAVG IGAVLFGFLG AAGSTMGAAS LTLTVQARQL LSGIVQQQSN LLRAIEAQQH LLRLTVWGIK QLQARVLAVE RYLSDQQLLG IWGCSGKLIC TTNVPWNSSW SNKSQDEIWN NMTWLQWDKE ISNYTDTIYY LIEKSQNQQE VNEKDLLALD KWTNLWNWFG ISNWLWYIRI FIMIVGGLIG LRIIFAVLSV INRVRQGYSP VSFQTLTPNP RELDRPGGIE EGDGELGKTR SIRLVGGFLA LFWDDLRSLC LFSYHRLRDF ILIAARILEL LGHNSLKGLR LGWEGLKYLG NLLLYWGREL KNSAVNLVDT IAIVVAGWTD RVIEVLQGIG RAFLHIPRRI RQGFERALL |
-Macromolecule #2: Immunoglobulin G PGT151 Fab, Heavy chain
| Macromolecule | Name: Immunoglobulin G PGT151 Fab, Heavy chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RVQLVESGGG VVQPGKSVRL SCVVSDFPFS KYPMYWVRQA PGKGLEWVAA ISGDAWHVVY SNSVQGRFLV SRDNVKNTLY LEMNSLKIE DTAVYRCARM FQESGPPRLD RWSGRNYYYY SGMDVWGQGT TVTVSSASTK GPSVFPLAPS SKSTSGGTAA L GCLVKDYF ...String: RVQLVESGGG VVQPGKSVRL SCVVSDFPFS KYPMYWVRQA PGKGLEWVAA ISGDAWHVVY SNSVQGRFLV SRDNVKNTLY LEMNSLKIE DTAVYRCARM FQESGPPRLD RWSGRNYYYY SGMDVWGQGT TVTVSSASTK GPSVFPLAPS SKSTSGGTAA L GCLVKDYF PEPVTVSWNS GALTSGVHTF PAVLQSSGLY SLSSVVTVPS SSLGTQTYIC NVNHKPSNTK VDKRVEPKSC DK |
-Macromolecule #3: Immunoglobulin G PGT151, Light chain
| Macromolecule | Name: Immunoglobulin G PGT151, Light chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQTPLS LSVTPGQPAS ISCKSSESLR QSNGKTSLYW YRQKPGQSPQ LLVFEVSNRF SGVSDRFVGS GSGTDFTLRI SRVEAEDVG FYYCMQSKDF PLTFGGGTKV DLKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV ...String: DIVMTQTPLS LSVTPGQPAS ISCKSSESLR QSNGKTSLYW YRQKPGQSPQ LLVFEVSNRF SGVSDRFVGS GSGTDFTLRI SRVEAEDVG FYYCMQSKDF PLTFGGGTKV DLKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV TEQDSKDSTY SLSSTLTLSK ADYEKHKVYA CEVTHQGLSS PVTKSFNRGE C |
-Macromolecule #4: Immunoglobulin G PCT64_35S Fab, Heavy chain
| Macromolecule | Name: Immunoglobulin G PCT64_35S Fab, Heavy chain / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVKPGGSLRL ACVGSEFTFS EAWMTWVRQA PGKGLEWVGH MRPTTEGGAK DYAAAVRGRF TIARDDSKST LYLQMNSLKI EDTGVYYCMT GVERGDFWSD DYSQHYNTYL IDVWGKGTTV TVSS |
-Macromolecule #5: Immunoglobulin G PCT64_35S, Light chain
| Macromolecule | Name: Immunoglobulin G PCT64_35S, Light chain / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGT LSLSPGDRAT LSCRATQSVG GDYFAWYQQR PGQSPRLLIY GTSRRAAGIP DRFSGSGSG TDFTLTIDRL EPEDFAVYYC RQYETSFTFG PGTKVDIK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 277.15 K / Instrument: HOMEMADE PLUNGER / Details: Specimen was manually frozen. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 980 / Average electron dose: 37.5 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)