[English] 日本語
 Yorodumi
Yorodumi- EMDB-21322: Full-length HIV-1 Envelope glycoprotein clone AMC011 in complex w... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21322 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Full-length HIV-1 Envelope glycoprotein clone AMC011 in complex with PGT151 Fab and VRC42.01 Fab | ||||||||||||
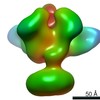
 Map data Map data | Full length HIV-1 Env AMC011 in complex with PGT151 Fab and VRC42.01 Fab | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.2 Å | ||||||||||||
 Authors Authors | Rantalainen K / Lee WS / Ward ABW | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2020 Journal: Cell Rep / Year: 2020Title: HIV-1 Envelope and MPER Antibody Structures in Lipid Assemblies. Authors: Kimmo Rantalainen / Zachary T Berndsen / Aleksandar Antanasijevic / Torben Schiffner / Xi Zhang / Wen-Hsin Lee / Jonathan L Torres / Lei Zhang / Adriana Irimia / Jeffrey Copps / Kenneth H ...Authors: Kimmo Rantalainen / Zachary T Berndsen / Aleksandar Antanasijevic / Torben Schiffner / Xi Zhang / Wen-Hsin Lee / Jonathan L Torres / Lei Zhang / Adriana Irimia / Jeffrey Copps / Kenneth H Zhou / Young D Kwon / William H Law / Chaim A Schramm / Raffaello Verardi / Shelly J Krebs / Peter D Kwong / Nicole A Doria-Rose / Ian A Wilson / Michael B Zwick / John R Yates / William R Schief / Andrew B Ward /  Abstract: Structural and functional studies of HIV envelope glycoprotein (Env) as a transmembrane protein have long been complicated by challenges associated with inherent flexibility of the molecule and the ...Structural and functional studies of HIV envelope glycoprotein (Env) as a transmembrane protein have long been complicated by challenges associated with inherent flexibility of the molecule and the membrane-embedded hydrophobic regions. Here, we present approaches for incorporating full-length, wild-type HIV-1 Env, as well as C-terminally truncated and stabilized versions, into lipid assemblies, providing a modular platform for Env structural studies by single particle electron microscopy. We reconstitute a full-length Env clone into a nanodisc, complex it with a membrane-proximal external region (MPER) targeting antibody 10E8, and structurally define the full quaternary epitope of 10E8 consisting of lipid, MPER, and ectodomain contacts. By aligning this and other Env-MPER antibody complex reconstructions with the lipid bilayer, we observe evidence of Env tilting as part of the neutralization mechanism for MPER-targeting antibodies. We also adapt the platform toward vaccine design purposes by introducing stabilizing mutations that allow purification of unliganded Env with a peptidisc scaffold. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21322.map.gz emd_21322.map.gz | 116.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21322-v30.xml emd-21322-v30.xml emd-21322.xml emd-21322.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21322_fsc.xml emd_21322_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_21322.png emd_21322.png | 166.4 KB | ||
| Others |  emd_21322_half_map_1.map.gz emd_21322_half_map_1.map.gz emd_21322_half_map_2.map.gz emd_21322_half_map_2.map.gz | 98.4 MB 98.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21322 http://ftp.pdbj.org/pub/emdb/structures/EMD-21322 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21322 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21322 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21322.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21322.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full length HIV-1 Env AMC011 in complex with PGT151 Fab and VRC42.01 Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Full length HIV-1 Env AMC011 in complex with...
| File | emd_21322_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full length HIV-1 Env AMC011 in complex with PGT151 Fab and VRC42.01 Fab half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Full length HIV-1 Env AMC011 in complex with...
| File | emd_21322_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full length HIV-1 Env AMC011 in complex with PGT151 Fab and VRC42.01 Fab half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Full-length HIV-1 Envelope glycoprotein clone AMC011 in complex w...
| Entire | Name: Full-length HIV-1 Envelope glycoprotein clone AMC011 in complex with PGT151 Fab and VRC42.01 Fab |
|---|---|
| Components |
|
-Supramolecule #1: Full-length HIV-1 Envelope glycoprotein clone AMC011 in complex w...
| Supramolecule | Name: Full-length HIV-1 Envelope glycoprotein clone AMC011 in complex with PGT151 Fab and VRC42.01 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#11 Details: AMC011 Env purified with stabilizing PGT151 Fab. Complexed with VRC42.01 Fab during detergent-lipid exchange and prior to grid preparation. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
| Details | AMC011 Env purified with stabilizing PGT151 Fab. Complexed with VRC42.01 Fab during detergent-lipid exchange and prior to grid preparation. Sample is in detergent-lipid micelle. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 3078 / Average electron dose: 51.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)