[English] 日本語
 Yorodumi
Yorodumi- EMDB-7858: Ectodomain of full length, wild type HIV-1 glycoprotein clone PC6... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7858 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ectodomain of full length, wild type HIV-1 glycoprotein clone PC64M18C043 in complex with PGT151 Fab | ||||||||||||
 Map data Map data | Ectodomain of full length, wild type HIV-1 glycoprotein clone PC64M18C043 in complex with PGT151 Fab | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | HIV-1 / Glycoprotein / Env / VIRAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationimmunoglobulin complex / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / adaptive immune response / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity ...immunoglobulin complex / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / adaptive immune response / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / extracellular region / metal ion binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | ||||||||||||
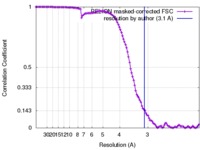
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | ||||||||||||
 Authors Authors | Berndsens ZB / Rantalainen KR / Ward AB | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2018 Journal: Cell Rep / Year: 2018Title: Co-evolution of HIV Envelope and Apex-Targeting Neutralizing Antibody Lineage Provides Benchmarks for Vaccine Design. Authors: Kimmo Rantalainen / Zachary T Berndsen / Sasha Murrell / Liwei Cao / Oluwarotimi Omorodion / Jonathan L Torres / Mengyu Wu / Jeffrey Umotoy / Jeffrey Copps / Pascal Poignard / Elise Landais ...Authors: Kimmo Rantalainen / Zachary T Berndsen / Sasha Murrell / Liwei Cao / Oluwarotimi Omorodion / Jonathan L Torres / Mengyu Wu / Jeffrey Umotoy / Jeffrey Copps / Pascal Poignard / Elise Landais / James C Paulson / Ian A Wilson / Andrew B Ward /  Abstract: Broadly neutralizing antibodies (bnAbs) targeting the HIV envelope glycoprotein (Env) typically take years to develop. Longitudinal analyses of both neutralizing antibody lineages and viruses at ...Broadly neutralizing antibodies (bnAbs) targeting the HIV envelope glycoprotein (Env) typically take years to develop. Longitudinal analyses of both neutralizing antibody lineages and viruses at serial time points during infection provide a basis for understanding the co-evolutionary contest between HIV and the humoral immune system. Here, we describe the structural characterization of an apex-targeting antibody lineage and autologous clade A viral Env from a donor in the Protocol C cohort. Comparison of Ab-Env complexes at early and late time points reveals that, within the antibody lineage, the CDRH3 loop rigidifies, the bnAb angle of approach steepens, and surface charges are mutated to accommodate glycan changes. Additionally, we observed differences in site-specific glycosylation between soluble and full-length Env constructs, which may be important for tuning optimal immunogenicity in soluble Env trimers. These studies therefore provide important guideposts for design of immunogens that prime and mature nAb responses to the Env V2-apex. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7858.map.gz emd_7858.map.gz | 117.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7858-v30.xml emd-7858-v30.xml emd-7858.xml emd-7858.xml | 22.4 KB 22.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7858_fsc.xml emd_7858_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_7858.png emd_7858.png | 382.1 KB | ||
| Masks |  emd_7858_msk_1.map emd_7858_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-7858.cif.gz emd-7858.cif.gz | 7.5 KB | ||
| Others |  emd_7858_half_map_1.map.gz emd_7858_half_map_1.map.gz emd_7858_half_map_2.map.gz emd_7858_half_map_2.map.gz | 98.2 MB 98.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7858 http://ftp.pdbj.org/pub/emdb/structures/EMD-7858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7858 | HTTPS FTP |
-Related structure data
| Related structure data |  6dcqMC  7859C  7860C  7861C  7862C  7863C  7864C  7865C  7866C  6ca6C  6ca7C  6ca9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7858.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7858.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ectodomain of full length, wild type HIV-1 glycoprotein clone PC64M18C043 in complex with PGT151 Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
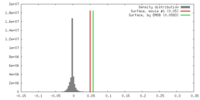
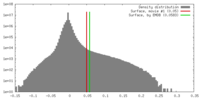
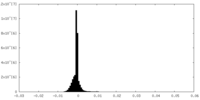
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_7858_msk_1.map emd_7858_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Ectodomain of full length, wild type HIV-1 glycoprotein...
| File | emd_7858_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ectodomain of full length, wild type HIV-1 glycoprotein clone PC64M18C043 in complex with PGT151 Fab | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Ectodomain of full length, wild type HIV-1 glycoprotein...
| File | emd_7858_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ectodomain of full length, wild type HIV-1 glycoprotein clone PC64M18C043 in complex with PGT151 Fab | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Full length, wild type HIV-1 Envelope glycoprotein in complex wit...
| Entire | Name: Full length, wild type HIV-1 Envelope glycoprotein in complex with PGT151 Fab |
|---|---|
| Components |
|
-Supramolecule #1: Full length, wild type HIV-1 Envelope glycoprotein in complex wit...
| Supramolecule | Name: Full length, wild type HIV-1 Envelope glycoprotein in complex with PGT151 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Full length HIV-1 Envelope glycoprotein clone C043 collected 18 months post infection from donor 64 of Protocol C cohort study. Expressed as recombinant protein in HEK293F cell line. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 389 KDa |
-Macromolecule #1: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 53.392359 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SAANNLWVTV YYGVPVWRDA ETTLFCASDA KAYDTEVHNV WATHACVPTD PSPQEIHLAN VTEKFDMWKN SMVEQMHTDI ISLWDESLK PCVKLTPLCI TLNCTNITRN VTGGNLTEEG KEELKNCSFN ATTELRDKIQ KVHSLFYRLD LVELNEGNSS D SNTSMYRL ...String: SAANNLWVTV YYGVPVWRDA ETTLFCASDA KAYDTEVHNV WATHACVPTD PSPQEIHLAN VTEKFDMWKN SMVEQMHTDI ISLWDESLK PCVKLTPLCI TLNCTNITRN VTGGNLTEEG KEELKNCSFN ATTELRDKIQ KVHSLFYRLD LVELNEGNSS D SNTSMYRL INCNTSAITQ ACPKVSFEPI PIHYCAPAGF AILKCREKEF NGTGPCKKVS TVQCTHGIKP VVSTQLLLNG SL AEGKVKI RCENISNNAK TILVQLTTPV RINCTRPSNN TRTSIRIGPG QSFYATGDII GDIRKAYCNV SESEWKEALG KVV EQLRNH FNKTITFASS SGGDLEITTH SFNCGGEFFY CNTSSLFNST WDGNSATNST QVPNGTITLP CRIKQIINMW QRTG QAMYA PPIPGKIRCD SNITGLILIR DGGNNNNESE TFRPGGGDMR NNWRSELYKY KVVKIDPLGV APTGAKRRVV EREKR UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 40.020207 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVLFG FLGAAGSTMG AASLTLTVQA RQLLSGIVQQ QSNLLRAIEA QQHLLRLTVW GIKQLQARVL AVERYLSDQQ LLGIWGCSG KLICTTNVPW NSSWSNKSQD EIWNNMTWLQ WDKEISNYTD TIYYLIEKSQ NQQEVNEKDL LALDKWTNLW N WFGISNWL ...String: AVGIGAVLFG FLGAAGSTMG AASLTLTVQA RQLLSGIVQQ QSNLLRAIEA QQHLLRLTVW GIKQLQARVL AVERYLSDQQ LLGIWGCSG KLICTTNVPW NSSWSNKSQD EIWNNMTWLQ WDKEISNYTD TIYYLIEKSQ NQQEVNEKDL LALDKWTNLW N WFGISNWL WYIRIFIMIV GGLIGLRIIF AVLSVINRVR QGYSPVSFQT LTPNPRELDR PGGIEEGDGE LGKTRSIRLV GG FLALFWD DLRSLCLFSY HRLRDFILIA ARILELLGHN SLKGLRLGWE GLKYLGNLLL YWGRELKNSA VNLVDTIAIV VAG WTDRVI EVLQGIGRAF LHIPRRIRQG FERALL UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: Immunoglobulin G PGT151 Fab, Heavy chain
| Macromolecule | Name: Immunoglobulin G PGT151 Fab, Heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.087438 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RVQLVESGGG VVQPGKSVRL SCVVSDFPFS KYPMYWVRQA PGKGLEWVAA ISGDAWHVVY SNSVQGRFLV SRDNVKNTLY LEMNSLKIE DTAVYRCARM FQESGPPRLD RWSGRNYYYY SGMDVWGQGT TVTVSSASTK GPSVFPLAPS SKSTSGGTAA L GCLVKDYF ...String: RVQLVESGGG VVQPGKSVRL SCVVSDFPFS KYPMYWVRQA PGKGLEWVAA ISGDAWHVVY SNSVQGRFLV SRDNVKNTLY LEMNSLKIE DTAVYRCARM FQESGPPRLD RWSGRNYYYY SGMDVWGQGT TVTVSSASTK GPSVFPLAPS SKSTSGGTAA L GCLVKDYF PEPVTVSWNS GALTSGVHTF PAVLQSSGLY SLSSVVTVPS SSLGTQTYIC NVNHKPSNTK VDKRVEPKSC DK UniProtKB: IgG H chain |
-Macromolecule #4: Immunoglobulin G PGT151 Fab, Light chain
| Macromolecule | Name: Immunoglobulin G PGT151 Fab, Light chain / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.057809 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQTPLS LSVTPGQPAS ISCKSSESLR QSNGKTSLYW YRQKPGQSPQ LLVFEVSNRF SGVSDRFVGS GSGTDFTLRI SRVEAEDVG FYYCMQSKDF PLTFGGGTKV DLKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV ...String: DIVMTQTPLS LSVTPGQPAS ISCKSSESLR QSNGKTSLYW YRQKPGQSPQ LLVFEVSNRF SGVSDRFVGS GSGTDFTLRI SRVEAEDVG FYYCMQSKDF PLTFGGGTKV DLKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV TEQDSKDSTY SLSSTLTLSK ADYEKHKVYA CEVTHQGLSS PVTKSFNRGE C UniProtKB: Ig-like domain-containing protein |
-Macromolecule #14: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 14 / Number of copies: 21 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: Detergent removed with Biobeads prior to grid freezing | |||||||||||||||
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: Blot force 0 Blot time 5 sec. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 4039 / Average electron dose: 1.885 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)