[English] 日本語
 Yorodumi
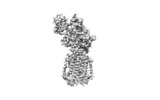
Yorodumi- EMDB-7796: Structure of human Patched1 in complex with native Sonic Hedgehog -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7796 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human Patched1 in complex with native Sonic Hedgehog | |||||||||
 Map data Map data | A protein structure | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of nodal signaling pathway / neural plate axis specification / response to chlorate / cell differentiation involved in kidney development / positive regulation of skeletal muscle cell proliferation / right lung development / left lung development / primary prostatic bud elongation / regulation of mesenchymal cell proliferation involved in prostate gland development / mesenchymal smoothened signaling pathway involved in prostate gland development ...regulation of nodal signaling pathway / neural plate axis specification / response to chlorate / cell differentiation involved in kidney development / positive regulation of skeletal muscle cell proliferation / right lung development / left lung development / primary prostatic bud elongation / regulation of mesenchymal cell proliferation involved in prostate gland development / mesenchymal smoothened signaling pathway involved in prostate gland development / positive regulation of sclerotome development / tracheoesophageal septum formation / negative regulation of ureter smooth muscle cell differentiation / positive regulation of ureter smooth muscle cell differentiation / negative regulation of kidney smooth muscle cell differentiation / positive regulation of kidney smooth muscle cell differentiation / morphogen activity / regulation of odontogenesis / positive regulation of mesenchymal cell proliferation involved in ureter development / hedgehog receptor activity / neural tube patterning / trunk neural crest cell migration / Formation of lateral plate mesoderm / cell proliferation involved in metanephros development / hindgut morphogenesis / polarity specification of anterior/posterior axis / negative regulation of alpha-beta T cell differentiation / regulation of prostatic bud formation / formation of anatomical boundary / smoothened binding / positive regulation of striated muscle cell differentiation / regulation of glial cell proliferation / metanephric mesenchymal cell proliferation involved in metanephros development / ventral midline development / trachea morphogenesis / hedgehog family protein binding / cholesterol-protein transferase activity / HHAT G278V doesn't palmitoylate Hh-Np / telencephalon regionalization / bud outgrowth involved in lung branching / epithelial-mesenchymal cell signaling / Ligand-receptor interactions / hindlimb morphogenesis / laminin-1 binding / lung epithelium development / salivary gland cavitation / negative regulation of cholesterol efflux / spinal cord dorsal/ventral patterning / negative regulation of mesenchymal cell apoptotic process / determination of left/right asymmetry in lateral mesoderm / epidermal cell fate specification / cell development / spinal cord motor neuron differentiation / negative regulation of T cell differentiation in thymus / establishment of epithelial cell polarity / positive regulation of T cell differentiation in thymus / skeletal muscle cell proliferation / prostate gland development / intermediate filament organization / limb bud formation / embryonic skeletal system development / stem cell development / skeletal muscle fiber differentiation / positive regulation of cerebellar granule cell precursor proliferation / limb morphogenesis / mesenchymal cell apoptotic process / animal organ formation / patched binding / embryonic digestive tract morphogenesis / negative regulation of cell division / hindbrain development / positive regulation of skeletal muscle tissue development / somite development / ectoderm development / embryonic foregut morphogenesis / negative regulation of dopaminergic neuron differentiation / epithelial cell proliferation involved in salivary gland morphogenesis / mesenchymal cell proliferation involved in lung development / neuron fate commitment / cerebellar granule cell precursor proliferation / Activation of SMO / self proteolysis / positive regulation of immature T cell proliferation in thymus / lung lobe morphogenesis / smooth muscle tissue development / dorsal/ventral neural tube patterning / artery development / lymphoid progenitor cell differentiation / positive regulation of astrocyte differentiation / CD4-positive or CD8-positive, alpha-beta T cell lineage commitment / pharyngeal system development / cellular response to cholesterol / regulation of stem cell proliferation / mammary gland duct morphogenesis / pattern specification process / mammary gland epithelial cell differentiation / negative thymic T cell selection / positive regulation of epithelial cell proliferation involved in prostate gland development / male genitalia development / branching involved in salivary gland morphogenesis Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / Resolution: 3.5 Å | |||||||||
 Authors Authors | Qi X / Li X | |||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Authors: Xiaofeng Qi / Philip Schmiege / Elias Coutavas / Jiawei Wang / Xiaochun Li /   Abstract: Hedgehog (HH) signalling governs embryogenesis and adult tissue homeostasis in mammals and other multicellular organisms. Whereas deficient HH signalling leads to birth defects, unrestrained HH ...Hedgehog (HH) signalling governs embryogenesis and adult tissue homeostasis in mammals and other multicellular organisms. Whereas deficient HH signalling leads to birth defects, unrestrained HH signalling is implicated in human cancers. N-terminally palmitoylated HH releases the repression of Patched to the oncoprotein smoothened (SMO); however, the mechanism by which HH recognizes Patched is unclear. Here we report cryo-electron microscopy structures of human patched 1 (PTCH1) alone and in complex with the N-terminal domain of 'native' sonic hedgehog (native SHH-N has both a C-terminal cholesterol and an N-terminal fatty-acid modification), at resolutions of 3.5 Å and 3.8 Å, respectively. The structure of PTCH1 has internal two-fold pseudosymmetry in the transmembrane core, which features a sterol-sensing domain and two homologous extracellular domains, resembling the architecture of Niemann-Pick C1 (NPC1) protein. The palmitoylated N terminus of SHH-N inserts into a cavity between the extracellular domains of PTCH1 and dominates the PTCH1-SHH-N interface, which is distinct from that reported for SHH-N co-receptors. Our biochemical assays show that SHH-N may use another interface, one that is required for its co-receptor binding, to recruit PTCH1 in the absence of a covalently attached palmitate. Our work provides atomic insights into the recognition of the N-terminal domain of HH (HH-N) by PTCH1, offers a structural basis for cooperative binding of HH-N to various receptors and serves as a molecular framework for HH signalling and its malfunction in disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7796.map.gz emd_7796.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7796-v30.xml emd-7796-v30.xml emd-7796.xml emd-7796.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7796.png emd_7796.png | 32.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7796 http://ftp.pdbj.org/pub/emdb/structures/EMD-7796 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7796 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7796 | HTTPS FTP |
-Validation report
| Summary document |  emd_7796_validation.pdf.gz emd_7796_validation.pdf.gz | 444.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7796_full_validation.pdf.gz emd_7796_full_validation.pdf.gz | 444.4 KB | Display | |
| Data in XML |  emd_7796_validation.xml.gz emd_7796_validation.xml.gz | 5.4 KB | Display | |
| Data in CIF |  emd_7796_validation.cif.gz emd_7796_validation.cif.gz | 6.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7796 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7796 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7796 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7796 | HTTPS FTP |
-Related structure data
| Related structure data |  6oevMC  7795C  6oeuC  6d4j C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7796.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7796.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A protein structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ptc
| Entire | Name: Ptc |
|---|---|
| Components |
|
-Supramolecule #1: Ptc
| Supramolecule | Name: Ptc / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Experimental: 121 KDa |
-Macromolecule #1: Protein patched homolog 1
| Macromolecule | Name: Protein patched homolog 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 160.714406 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASAGNAAEP QDRGGGGSGC IGAPGRPAGG GRRRRTGGLR RAAAPDRDYL HRPSYCDAAF ALEQISKGKA TGRKAPLWLR AKFQRLLFK LGCYIQKNCG KFLVVGLLIF GAFAVGLKAA NLETNVEELW VEVGGRVSRE LNYTRQKIGE EAMFNPQLMI Q TPKEEGAN ...String: MASAGNAAEP QDRGGGGSGC IGAPGRPAGG GRRRRTGGLR RAAAPDRDYL HRPSYCDAAF ALEQISKGKA TGRKAPLWLR AKFQRLLFK LGCYIQKNCG KFLVVGLLIF GAFAVGLKAA NLETNVEELW VEVGGRVSRE LNYTRQKIGE EAMFNPQLMI Q TPKEEGAN VLTTEALLQH LDSALQASRV HVYMYNRQWK LEHLCYKSGE LITETGYMDQ IIEYLYPCLI ITPLDCFWEG AK LQSGTAY LLGKPPLRWT NFDPLEFLEE LKKINYQVDS WEEMLNKAEV GHGYMDRPCL NPADPDCPAT APNKNSTKPL DMA LVLNGG CHGLSRKYMH WQEELIVGGT VKNSTGKLVS AHALQTMFQL MTPKQMYEHF KGYEYVSHIN WNEDKAAAIL EAWQ RTYVE VVHQSVAQNS TQKVLSFTTT TLDDILKSFS DVSVIRVASG YLLMLAYACL TMLRWDCSKS QGAVGLAGVL LVALS VAAG LGLCSLIGIS FNAATTQVLP FLALGVGVDD VFLLAHAFSE TGQNKRIPFE DRTGECLKRT GASVALTSIS NVTAFF MAA LIPIPALRAF SLQAAVVVVF NFAMVLLIFP AILSMDLYRR EDRRLDIFCC FTSPCVSRVI QVEPQAYTDT HDNTRYS PP PPYSSHSFAH ETQITMQSTV QLRTEYDPHT HVYYTTAEPR SEISVQPVTV TQDTLSCQSP ESTSSTRDLL SQFSDSSL H CLEPPCTKWT LSSFAEKHYA PFLLKPKAKV VVIFLFLGLL GVSLYGTTRV RDGLDLTDIV PRETREYDFI AAQFKYFSF YNMYIVTQKA DYPNIQHLLY DLHRSFSNVK YVMLEENKQL PKMWLHYFRD WLQGLQDAFD SDWETGKIMP NNYKNGSDDG VLAYKLLVQ TGSRDKPIDI SQLTKQRLVD ADGIINPSAF YIYLTAWVSN DPVAYAASQA NIRPHRPEWV HDKADYMPET R LRIPAAEP IEYAQFPFYL NGLRDTSDFV EAIEKVRTIC SNYTSLGLSS YPNGYPFLFW EQYIGLRHWL LLFISVVLAC TF LVCAVFL LNPWTAGIIV MVLALMTVEL FGMMGLIGIK LSAVPVVILI ASVGIGVEFT VHVALAFLTA IGDKNRRAVL ALE HMFAPV LDGAVSTLLG VLMLAGSEFD FIVRYFFAVL AILTILGVLN GLVLLPVLLS FFGPYPEVSP ANGLNRLPTP SPEP PPSVV RFAMPPGHTH SGSDSSDSEY SSQTTVSGLS EELRHYEAQQ GAGGPAHQVI VEATENPVFA HSTVVHPESR HHPPS NPRQ QPHLDSGSLP PGRQGQQPRR DPPREGLWPP PYRPRRDAFE ISTEGHSGPS NRARWGPRGA RSHNPRNPAS TAMGSS VPG YCQPITTVTA SASVTVAVHP PPVPGPGRNP RGGLCPGYPE TDHGLFEDPH VPFHVRCERR DSKVEVIELQ DVECEER PR GSSSN |
-Macromolecule #2: Sonic hedgehog protein
| Macromolecule | Name: Sonic hedgehog protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 19.594039 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: CGPGRGFGKR RHPKKLTPLA YKQFIPNVAE KTLGASGRYE GKISRNSERF KELTPNYNPD IIFKDEENTG ADRLMTQRCK DKLNALAIS VMNQWPGVKL RVTEGWDEDG HHSEESLHYE GRAVDITTSD RDRSKYGMLA RLAVEAGFDW VYYESKAHIH C SVKAENSV AAKSGG |
-Macromolecule #3: N-ACETYL-D-GLUCOSAMINE
| Macromolecule | Name: N-ACETYL-D-GLUCOSAMINE / type: ligand / ID: 3 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #5: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 5 / Number of copies: 1 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Experimental details
-Structure determination
 Processing Processing | single particle reconstruction |
|---|---|
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 789118 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)