+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22074 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of NLRP1-DPP9 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NLRP1 / DPP9 / inflammasome / Val-boroPro (VbP) / talabostat / innate immunity / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationNLRP1 inflammasome complex / NLRP1 inflammasome complex assembly / The NLRP1 inflammasome / self proteolysis / NLRP3 inflammasome complex / dipeptidyl-peptidase IV / cysteine-type endopeptidase activator activity / dipeptidyl-peptidase activity / Hydrolases; Acting on peptide bonds (peptidases) / negative regulation of programmed cell death ...NLRP1 inflammasome complex / NLRP1 inflammasome complex assembly / The NLRP1 inflammasome / self proteolysis / NLRP3 inflammasome complex / dipeptidyl-peptidase IV / cysteine-type endopeptidase activator activity / dipeptidyl-peptidase activity / Hydrolases; Acting on peptide bonds (peptidases) / negative regulation of programmed cell death / pattern recognition receptor signaling pathway / cellular response to UV-B / cysteine-type endopeptidase activator activity involved in apoptotic process / pattern recognition receptor activity / response to muramyl dipeptide / cell leading edge / pyroptotic inflammatory response / aminopeptidase activity / signaling adaptor activity / antiviral innate immune response / activation of innate immune response / intrinsic apoptotic signaling pathway / serine-type peptidase activity / positive regulation of interleukin-1 beta production / molecular condensate scaffold activity / protein homooligomerization / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / positive regulation of inflammatory response / peptidase activity / double-stranded RNA binding / regulation of inflammatory response / neuron apoptotic process / double-stranded DNA binding / regulation of apoptotic process / defense response to virus / microtubule / defense response to bacterium / inflammatory response / protein domain specific binding / apoptotic process / nucleolus / enzyme binding / signal transduction / ATP hydrolysis activity / proteolysis / nucleoplasm / ATP binding / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Hollingsworth LR / Sharif H / Griswold AR / Fontana P / Mintseris J / Dagbay KB / Paulo JA / Gygi SP / Bachovchin DA / Wu H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation. Authors: L Robert Hollingsworth / Humayun Sharif / Andrew R Griswold / Pietro Fontana / Julian Mintseris / Kevin B Dagbay / Joao A Paulo / Steven P Gygi / Daniel A Bachovchin / Hao Wu /  Abstract: Nucleotide-binding domain and leucine-rich repeat pyrin-domain containing protein 1 (NLRP1) is an inflammasome sensor that mediates the activation of caspase-1 to induce cytokine maturation and ...Nucleotide-binding domain and leucine-rich repeat pyrin-domain containing protein 1 (NLRP1) is an inflammasome sensor that mediates the activation of caspase-1 to induce cytokine maturation and pyroptosis. Gain-of-function mutations of NLRP1 cause severe inflammatory diseases of the skin. NLRP1 contains a function-to-find domain that auto-proteolyses into noncovalently associated subdomains, and proteasomal degradation of the repressive N-terminal fragment of NLRP1 releases its inflammatory C-terminal fragment (NLRP1 CT). Cytosolic dipeptidyl peptidases 8 and 9 (hereafter, DPP8/DPP9) both interact with NLRP1, and small-molecule inhibitors of DPP8/DPP9 activate NLRP1 by mechanisms that are currently unclear. Here we report cryo-electron microscopy structures of the human NLRP1-DPP9 complex alone and with Val-boroPro (VbP), an inhibitor of DPP8/DPP9. The structures reveal a ternary complex that comprises DPP9, full-length NLRP1 and the NLRPT CT. The binding of the NLRP1 CT to DPP9 requires full-length NLRP1, which suggests that NLRP1 activation is regulated by the ratio of NLRP1 CT to full-length NLRP1. Activation of the inflammasome by ectopic expression of the NLRP1 CT is consistently rescued by co-expression of autoproteolysis-deficient full-length NLRP1. The N terminus of the NLRP1 CT inserts into the DPP9 active site, and VbP disrupts this interaction. Thus, VbP weakens the NLRP1-DPP9 interaction and accelerates degradation of the N-terminal fragment to induce inflammasome activation. Overall, these data demonstrate that DPP9 quenches low levels of NLRP1 CT and thus serves as a checkpoint for activation of the NLRP1 inflammasome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22074.map.gz emd_22074.map.gz | 15.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22074-v30.xml emd-22074-v30.xml emd-22074.xml emd-22074.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22074_fsc.xml emd_22074_fsc.xml | 14.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_22074.png emd_22074.png | 161.7 KB | ||
| Filedesc metadata |  emd-22074.cif.gz emd-22074.cif.gz | 7.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22074 http://ftp.pdbj.org/pub/emdb/structures/EMD-22074 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22074 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22074 | HTTPS FTP |
-Related structure data
| Related structure data |  6x6aMC  6x6cC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10594 (Title: Human NLRP1-DPP9 complex / Data size: 2.3 TB EMPIAR-10594 (Title: Human NLRP1-DPP9 complex / Data size: 2.3 TBData #1: Unaligned multi frame micographs of NLRP1-DPP9-Apo-noTILT dataset [micrographs - multiframe] Data #2: Unaligned multi frame micographs of NLRP1-DPP9-Apo-TILT dataset [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22074.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22074.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
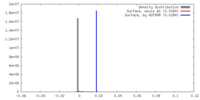
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DPP9-NLRP1 complex
| Entire | Name: DPP9-NLRP1 complex |
|---|---|
| Components |
|
-Supramolecule #1: DPP9-NLRP1 complex
| Supramolecule | Name: DPP9-NLRP1 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Dipeptidyl peptidase 9
| Macromolecule | Name: Dipeptidyl peptidase 9 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: dipeptidyl-peptidase IV |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 98.38432 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MATTGTPTAD RGDAAATDDP AARFQVQKHS WDGLRSIIHG SRKYSGLIVN KAPHDFQFVQ KTDESGPHSH RLYYLGMPYG SRENSLLYS EIPKKVRKEA LLLLSWKQML DHFQATPHHG VYSREEELLR ERKRLGVFGI TSYDFHSESG LFLFQASNSL F HCRDGGKN ...String: MATTGTPTAD RGDAAATDDP AARFQVQKHS WDGLRSIIHG SRKYSGLIVN KAPHDFQFVQ KTDESGPHSH RLYYLGMPYG SRENSLLYS EIPKKVRKEA LLLLSWKQML DHFQATPHHG VYSREEELLR ERKRLGVFGI TSYDFHSESG LFLFQASNSL F HCRDGGKN GFMVSPMKPL EIKTQCSGPR MDPKICPADP AFFSFINNSD LWVANIETGE ERRLTFCHQG LSNVLDDPKS AG VATFVIQ EEFDRFTGYW WCPTASWEGS EGLKTLRILY EEVDESEVEV IHVPSPALEE RKTDSYRYPR TGSKNPKIAL KLA EFQTDS QGKIVSTQEK ELVQPFSSLF PKVEYIARAG WTRDGKYAWA MFLDRPQQWL QLVLLPPALF IPSTENEEQR LASA RAVPR NVQPYVVYEE VTNVWINVHD IFYPFPQSEG EDELCFLRAN ECKTGFCHLY KVTAVLKSQG YDWSEPFSPG EDEFK CPIK EEIALTSGEW EVLARHGSKI WVNEETKLVY FQGTKDTPLE HHLYVVSYEA AGEIVRLTTP GFSHSCSMSQ NFDMFV SHY SSVSTPPCVH VYKLSGPDDD PLHKQPRFWA SMMEAASCPP DYVPPEIFHF HTRSDVRLYG MIYKPHALQP GKKHPTV LF VYGGPQVQLV NNSFKGIKYL RLNTLASLGY AVVVIDGRGS CQRGLRFEGA LKNQMGQVEI EDQVEGLQFV AEKYGFID L SRVAIHGWSY GGFLSLMGLI HKPQVFKVAI AGAPVTVWMA YDTGYTERYM DVPENNQHGY EAGSVALHVE KLPNEPNRL LILHGFLDEN VHFFHTNFLV SQLIRAGKPY QLQIYPNERH SIRCPESGEH YEVTLLHFLQ EYL UniProtKB: Dipeptidyl peptidase 9 |
-Macromolecule #2: NACHT, LRR and PYD domains-containing protein 1
| Macromolecule | Name: NACHT, LRR and PYD domains-containing protein 1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 136.327344 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAGGAWGRLA CYLEFLKKEE LKEFQLLLAN KAHSRSSSGE TPAQPEKTSG MEVASYLVAQ YGEQRAWDLA LHTWEQMGLR SLCAQAQEG AGHSPSFPYS PSEPHLGSPS QPTSTAVLMP WIHELPAGCT QGSERRVLRQ LPDTSGRRWR EISASLLYQA L PSSPDHES ...String: MAGGAWGRLA CYLEFLKKEE LKEFQLLLAN KAHSRSSSGE TPAQPEKTSG MEVASYLVAQ YGEQRAWDLA LHTWEQMGLR SLCAQAQEG AGHSPSFPYS PSEPHLGSPS QPTSTAVLMP WIHELPAGCT QGSERRVLRQ LPDTSGRRWR EISASLLYQA L PSSPDHES PSQESPNAPT STAVLGSWGS PPQPSLAPRE QEAPGTQWPL DETSGIYYTE IREREREKSE KGRPPWAAVV GT PPQAHTS LQPHHHPWEP SVRESLCSTW PWKNEDFNQK FTQLLLLQRP HPRSQDPLVK RSWPDYVEEN RGHLIEIRDL FGP GLDTQE PRIVILQGAA GIGKSTLARQ VKEAWGRGQL YGDRFQHVFY FSCRELAQSK VVSLAELIGK DGTATPAPIR QILS RPERL LFILDGVDEP GWVLQEPSSE LCLHWSQPQP ADALLGSLLG KTILPEASFL ITARTTALQN LIPSLEQARW VEVLG FSES SRKEYFYRYF TDERQAIRAF RLVKSNKELW ALCLVPWVSW LACTCLMQQM KRKEKLTLTS KTTTTLCLHY LAQALQ AQP LGPQLRDLCS LAAEGIWQKK TLFSPDDLRK HGLDGAIIST FLKMGILQEH PIPLSYSFIH LCFQEFFAAM SYVLEDE KG RGKHSNCIID LEKTLEAYGI HGLFGASTTR FLLGLLSDEG EREMENIFHC RLSQGRNLMQ WVPSLQLLLQ PHSLESLH C LYETRNKTFL TQVMAHFEEM GMCVETDMEL LVCTFCIKFS RHVKKLQLIE GRQHRSTWSP TMVVLFRWVP VTDAYWQIL FSVLKVTRNL KELDLSGNSL SHSAVKSLCK TLRRPRCLLE TLRLAGCGLT AEDCKDLAFG LRANQTLTEL DLSFNVLTDA GAKHLCQRL RQPSCKLQRL QLVSCGLTSD CCQDLASVLS ASPSLKELDL QQNNLDDVGV RLLCEGLRHP ACKLIRLGLD Q TTLSDEMR QELRALEQEK PQLLIFSRRK PSVMTPTEGL DTGEMSNSTS SLKRQRLGSE RAASHVAQAN LKLLDVSKIF PI AEIAEES SPEVVPVELL CVPSPASQGD LHTKPLGTDD DFWGPTGPVA TEVVDKEKNL YRVHFPVAGS YRWPNTGLCF VMR EAVTVE IEFCVWDQFL GEINPQHSWM VAGPLLDIKA EPGAVEAVHL PHFVALQGGH VDTSLFQMAH FKEEGMLLEK PARV ELHHI VLENPSF UniProtKB: NACHT, LRR and PYD domains-containing protein 1 |
-Macromolecule #3: NACHT, LRR and PYD domains-containing protein 1
| Macromolecule | Name: NACHT, LRR and PYD domains-containing protein 1 / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.76074 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SPLGVLLKMI HNALRFIPVT SVVLLYHRVH PEEVTFHLYL IPSDCSIRKA IDDLEMKFQF VRIHKPPPLT PLYMGCRYTV SGSGSGMLE ILPKELELCY RSPGEDQLFS EFYVGHLGSG IRLQVKDKKD ETLVWEALVK PGDLMPATTL IPPARIAVPS P LDAPQLLH ...String: SPLGVLLKMI HNALRFIPVT SVVLLYHRVH PEEVTFHLYL IPSDCSIRKA IDDLEMKFQF VRIHKPPPLT PLYMGCRYTV SGSGSGMLE ILPKELELCY RSPGEDQLFS EFYVGHLGSG IRLQVKDKKD ETLVWEALVK PGDLMPATTL IPPARIAVPS P LDAPQLLH FVDQYREQLI ARVTSVEVVL DKLHGQVLSQ EQYERVLAEN TRPSQMRKLF SLSQSWDRKC KDGLYQALKE TH PHLIMEL WEKGSKKGLL PLSS UniProtKB: NACHT, LRR and PYD domains-containing protein 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 25 mM Tris pH 7.5, 150 mM NaCl, 1 mM TCEP |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / #0 - Number grids imaged: 7 / #0 - Number real images: 7840 / #0 - Average exposure time: 1.8 sec. / #0 - Average electron dose: 67.54 e/Å2 / #0 - Details: stage tilt 0 degrees / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / #1 - Number grids imaged: 4 / #1 - Number real images: 1916 / #1 - Average exposure time: 2.6 sec. / #1 - Average electron dose: 67.6 e/Å2 / #1 - Details: stage tilt 37 degrees |
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Calibrated magnification: 10500 / Illumination mode: SPOT SCAN / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: -1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera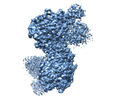



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)