+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22075 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of NLRP1-DPP9-VbP complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NLRP1 / DPP9 / inflammasome / Val-boroPro (VbP) / talabostat / innate immunity / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationNLRP1 inflammasome complex / NLRP1 inflammasome complex assembly / The NLRP1 inflammasome / self proteolysis / NLRP3 inflammasome complex / dipeptidyl-peptidase IV / cysteine-type endopeptidase activator activity / dipeptidyl-peptidase activity / Hydrolases; Acting on peptide bonds (peptidases) / negative regulation of programmed cell death ...NLRP1 inflammasome complex / NLRP1 inflammasome complex assembly / The NLRP1 inflammasome / self proteolysis / NLRP3 inflammasome complex / dipeptidyl-peptidase IV / cysteine-type endopeptidase activator activity / dipeptidyl-peptidase activity / Hydrolases; Acting on peptide bonds (peptidases) / negative regulation of programmed cell death / pattern recognition receptor signaling pathway / cellular response to UV-B / cysteine-type endopeptidase activator activity involved in apoptotic process / pattern recognition receptor activity / response to muramyl dipeptide / cell leading edge / pyroptotic inflammatory response / aminopeptidase activity / signaling adaptor activity / antiviral innate immune response / activation of innate immune response / intrinsic apoptotic signaling pathway / serine-type peptidase activity / positive regulation of interleukin-1 beta production / molecular condensate scaffold activity / protein homooligomerization / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / positive regulation of inflammatory response / peptidase activity / double-stranded RNA binding / regulation of inflammatory response / neuron apoptotic process / double-stranded DNA binding / regulation of apoptotic process / defense response to virus / microtubule / defense response to bacterium / inflammatory response / protein domain specific binding / apoptotic process / nucleolus / enzyme binding / signal transduction / ATP hydrolysis activity / proteolysis / nucleoplasm / ATP binding / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
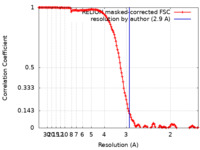
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Hollingsworth LR / Sharif H / Griswold AR / Fontana P / Mintseris J / Dagbay KB / Paulo JA / Gygi SP / Bachovchin DA / Wu H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation. Authors: L Robert Hollingsworth / Humayun Sharif / Andrew R Griswold / Pietro Fontana / Julian Mintseris / Kevin B Dagbay / Joao A Paulo / Steven P Gygi / Daniel A Bachovchin / Hao Wu /  Abstract: Nucleotide-binding domain and leucine-rich repeat pyrin-domain containing protein 1 (NLRP1) is an inflammasome sensor that mediates the activation of caspase-1 to induce cytokine maturation and ...Nucleotide-binding domain and leucine-rich repeat pyrin-domain containing protein 1 (NLRP1) is an inflammasome sensor that mediates the activation of caspase-1 to induce cytokine maturation and pyroptosis. Gain-of-function mutations of NLRP1 cause severe inflammatory diseases of the skin. NLRP1 contains a function-to-find domain that auto-proteolyses into noncovalently associated subdomains, and proteasomal degradation of the repressive N-terminal fragment of NLRP1 releases its inflammatory C-terminal fragment (NLRP1 CT). Cytosolic dipeptidyl peptidases 8 and 9 (hereafter, DPP8/DPP9) both interact with NLRP1, and small-molecule inhibitors of DPP8/DPP9 activate NLRP1 by mechanisms that are currently unclear. Here we report cryo-electron microscopy structures of the human NLRP1-DPP9 complex alone and with Val-boroPro (VbP), an inhibitor of DPP8/DPP9. The structures reveal a ternary complex that comprises DPP9, full-length NLRP1 and the NLRPT CT. The binding of the NLRP1 CT to DPP9 requires full-length NLRP1, which suggests that NLRP1 activation is regulated by the ratio of NLRP1 CT to full-length NLRP1. Activation of the inflammasome by ectopic expression of the NLRP1 CT is consistently rescued by co-expression of autoproteolysis-deficient full-length NLRP1. The N terminus of the NLRP1 CT inserts into the DPP9 active site, and VbP disrupts this interaction. Thus, VbP weakens the NLRP1-DPP9 interaction and accelerates degradation of the N-terminal fragment to induce inflammasome activation. Overall, these data demonstrate that DPP9 quenches low levels of NLRP1 CT and thus serves as a checkpoint for activation of the NLRP1 inflammasome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22075.map.gz emd_22075.map.gz | 14.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22075-v30.xml emd-22075-v30.xml emd-22075.xml emd-22075.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22075_fsc.xml emd_22075_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_22075.png emd_22075.png | 150.9 KB | ||
| Filedesc metadata |  emd-22075.cif.gz emd-22075.cif.gz | 8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22075 http://ftp.pdbj.org/pub/emdb/structures/EMD-22075 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22075 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22075 | HTTPS FTP |
-Related structure data
| Related structure data |  6x6cMC  6x6aC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10595 (Title: Human NLRP1-DPP9-VbP complex / Data size: 1.2 TB EMPIAR-10595 (Title: Human NLRP1-DPP9-VbP complex / Data size: 1.2 TBData #1: Unaligned multi frame micographs of NLRP1-DPP9-VbP-noTILT [micrographs - focal pairs - unprocessed] Data #2: Unaligned multi frame micographs of NLRP1-DPP9-VbP-TILT [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22075.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22075.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DPP9-NLRP1 complex
| Entire | Name: DPP9-NLRP1 complex |
|---|---|
| Components |
|
-Supramolecule #1: DPP9-NLRP1 complex
| Supramolecule | Name: DPP9-NLRP1 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Dipeptidyl peptidase 9
| Macromolecule | Name: Dipeptidyl peptidase 9 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: dipeptidyl-peptidase IV |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 101.761984 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSYYHHHHHH DYDIPTTENL YFQGAMGSMA TTGTPTADRG DAAATDDPAA RFQVQKHSWD GLRSIIHGSR KYSGLIVNKA PHDFQFVQK TDESGPHSHR LYYLGMPYGS RENSLLYSEI PKKVRKEALL LLSWKQMLDH FQATPHHGVY SREEELLRER K RLGVFGIT ...String: MSYYHHHHHH DYDIPTTENL YFQGAMGSMA TTGTPTADRG DAAATDDPAA RFQVQKHSWD GLRSIIHGSR KYSGLIVNKA PHDFQFVQK TDESGPHSHR LYYLGMPYGS RENSLLYSEI PKKVRKEALL LLSWKQMLDH FQATPHHGVY SREEELLRER K RLGVFGIT SYDFHSESGL FLFQASNSLF HCRDGGKNGF MVSPMKPLEI KTQCSGPRMD PKICPADPAF FSFINNSDLW VA NIETGEE RRLTFCHQGL SNVLDDPKSA GVATFVIQEE FDRFTGYWWC PTASWEGSEG LKTLRILYEE VDESEVEVIH VPS PALEER KTDSYRYPRT GSKNPKIALK LAEFQTDSQG KIVSTQEKEL VQPFSSLFPK VEYIARAGWT RDGKYAWAMF LDRP QQWLQ LVLLPPALFI PSTENEEQRL ASARAVPRNV QPYVVYEEVT NVWINVHDIF YPFPQSEGED ELCFLRANEC KTGFC HLYK VTAVLKSQGY DWSEPFSPGE DEFKCPIKEE IALTSGEWEV LARHGSKIWV NEETKLVYFQ GTKDTPLEHH LYVVSY EAA GEIVRLTTPG FSHSCSMSQN FDMFVSHYSS VSTPPCVHVY KLSGPDDDPL HKQPRFWASM MEAASCPPDY VPPEIFH FH TRSDVRLYGM IYKPHALQPG KKHPTVLFVY GGPQVQLVNN SFKGIKYLRL NTLASLGYAV VVIDGRGSCQ RGLRFEGA L KNQMGQVEIE DQVEGLQFVA EKYGFIDLSR VAIHGWSYGG FLSLMGLIHK PQVFKVAIAG APVTVWMAYD TGYTERYMD VPENNQHGYE AGSVALHVEK LPNEPNRLLI LHGFLDENVH FFHTNFLVSQ LIRAGKPYQL QIYPNERHSI RCPESGEHYE VTLLHFLQE YL UniProtKB: Dipeptidyl peptidase 9 |
-Macromolecule #2: NACHT, LRR and PYD domains-containing protein 1
| Macromolecule | Name: NACHT, LRR and PYD domains-containing protein 1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 166.069484 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAGGAWGRLA CYLEFLKKEE LKEFQLLLAN KAHSRSSSGE TPAQPEKTSG MEVASYLVAQ YGEQRAWDLA LHTWEQMGLR SLCAQAQEG AGHSPSFPYS PSEPHLGSPS QPTSTAVLMP WIHELPAGCT QGSERRVLRQ LPDTSGRRWR EISASLLYQA L PSSPDHES ...String: MAGGAWGRLA CYLEFLKKEE LKEFQLLLAN KAHSRSSSGE TPAQPEKTSG MEVASYLVAQ YGEQRAWDLA LHTWEQMGLR SLCAQAQEG AGHSPSFPYS PSEPHLGSPS QPTSTAVLMP WIHELPAGCT QGSERRVLRQ LPDTSGRRWR EISASLLYQA L PSSPDHES PSQESPNAPT STAVLGSWGS PPQPSLAPRE QEAPGTQWPL DETSGIYYTE IREREREKSE KGRPPWAAVV GT PPQAHTS LQPHHHPWEP SVRESLCSTW PWKNEDFNQK FTQLLLLQRP HPRSQDPLVK RSWPDYVEEN RGHLIEIRDL FGP GLDTQE PRIVILQGAA GIGKSTLARQ VKEAWGRGQL YGDRFQHVFY FSCRELAQSK VVSLAELIGK DGTATPAPIR QILS RPERL LFILDGVDEP GWVLQEPSSE LCLHWSQPQP ADALLGSLLG KTILPEASFL ITARTTALQN LIPSLEQARW VEVLG FSES SRKEYFYRYF TDERQAIRAF RLVKSNKELW ALCLVPWVSW LACTCLMQQM KRKEKLTLTS KTTTTLCLHY LAQALQ AQP LGPQLRDLCS LAAEGIWQKK TLFSPDDLRK HGLDGAIIST FLKMGILQEH PIPLSYSFIH LCFQEFFAAM SYVLEDE KG RGKHSNCIID LEKTLEAYGI HGLFGASTTR FLLGLLSDEG EREMENIFHC RLSQGRNLMQ WVPSLQLLLQ PHSLESLH C LYETRNKTFL TQVMAHFEEM GMCVETDMEL LVCTFCIKFS RHVKKLQLIE GRQHRSTWSP TMVVLFRWVP VTDAYWQIL FSVLKVTRNL KELDLSGNSL SHSAVKSLCK TLRRPRCLLE TLRLAGCGLT AEDCKDLAFG LRANQTLTEL DLSFNVLTDA GAKHLCQRL RQPSCKLQRL QLVSCGLTSD CCQDLASVLS ASPSLKELDL QQNNLDDVGV RLLCEGLRHP ACKLIRLGLD Q TTLSDEMR QELRALEQEK PQLLIFSRRK PSVMTPTEGL DTGEMSNSTS SLKRQRLGSE RAASHVAQAN LKLLDVSKIF PI AEIAEES SPEVVPVELL CVPSPASQGD LHTKPLGTDD DFWGPTGPVA TEVVDKEKNL YRVHFPVAGS YRWPNTGLCF VMR EAVTVE IEFCVWDQFL GEINPQHSWM VAGPLLDIKA EPGAVEAVHL PHFVALQGGH VDTSLFQMAH FKEEGMLLEK PARV ELHHI VLENPSFSPL GVLLKMIHNA LRFIPVTSVV LLYHRVHPEE VTFHLYLIPS DCSIRKAIDD LEMKFQFVRI HKPPP LTPL YMGCRYTVSG SGSGMLEILP KELELCYRSP GEDQLFSEFY VGHLGSGIRL QVKDKKDETL VWEALVKPGD LMPATT LIP PARIAVPSPL DAPQLLHFVD QYREQLIARV TSVEVVLDKL HGQVLSQEQY ERVLAENTRP SQMRKLFSLS QSWDRKC KD GLYQALKETH PHLIMELWEK GSKKGLLPLS S UniProtKB: NACHT, LRR and PYD domains-containing protein 1 |
-Macromolecule #3: [(2~{R})-1-[(2~{R})-2-azanyl-3-methyl-butanoyl]pyrrolidin-2-yl]bo...
| Macromolecule | Name: [(2~{R})-1-[(2~{R})-2-azanyl-3-methyl-butanoyl]pyrrolidin-2-yl]boronic acid type: ligand / ID: 3 / Number of copies: 2 / Formula: GK2 |
|---|---|
| Molecular weight | Theoretical: 214.07 Da |
| Chemical component information | 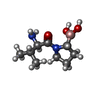 ChemComp-GK2: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 25 mM Tris pH 7.5, 150 mM NaCl, 1 mM TCEP |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / #0 - Number grids imaged: 4 / #0 - Number real images: 3553 / #0 - Average exposure time: 2.0 sec. / #0 - Average electron dose: 52.0 e/Å2 / #0 - Details: stage tilt 0 degrees / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / #1 - Number grids imaged: 4 / #1 - Number real images: 1954 / #1 - Average exposure time: 2.4 sec. / #1 - Average electron dose: 65.0 e/Å2 / #1 - Details: stage tilt 37 degrees |
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Calibrated magnification: 10500 / Illumination mode: SPOT SCAN / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: -0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)