[English] 日本語
 Yorodumi
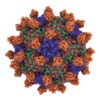
Yorodumi- EMDB-7772: Cryo-EM structure of Seneca Valley Virus-Anthrax Toxin Receptor 1... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7772 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Seneca Valley Virus-Anthrax Toxin Receptor 1 complex | |||||||||
 Map data Map data | Seneca Valley Virus-Anthrax Toxin Receptor 1 complex | |||||||||
 Sample Sample | Seneca Valley Virus-Anthrax Toxin Receptor 1 complex != Senecavirus A Seneca Valley Virus-Anthrax Toxin Receptor 1 complex
| |||||||||
 Keywords Keywords | Virus-receptor complex / Picornavirus / Senecavirus / Anthrax Toxin Receptor / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF7 activity / host cell nucleolus / adhesion receptor-mediated virion attachment to host cell / filopodium membrane / negative regulation of extracellular matrix assembly / symbiont-mediated suppression of host TRAF-mediated signal transduction / blood vessel development / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / lamellipodium membrane / Uptake and function of anthrax toxins ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF7 activity / host cell nucleolus / adhesion receptor-mediated virion attachment to host cell / filopodium membrane / negative regulation of extracellular matrix assembly / symbiont-mediated suppression of host TRAF-mediated signal transduction / blood vessel development / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / lamellipodium membrane / Uptake and function of anthrax toxins / collagen binding / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / substrate adhesion-dependent cell spreading / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / actin filament binding / transmembrane signaling receptor activity / channel activity / actin cytoskeleton organization / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / monoatomic ion transmembrane transport / symbiont-mediated suppression of host toll-like receptor signaling pathway / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity / entry receptor-mediated virion attachment to host cell / ubiquitinyl hydrolase 1 / cysteine-type deubiquitinase activity / RNA helicase activity / endosome membrane / RNA helicase / external side of plasma membrane / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / symbiont entry into host cell / DNA-templated transcription / virion attachment to host cell / structural molecule activity / cell surface / ATP hydrolysis activity / proteolysis / RNA binding / ATP binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Senecavirus A Senecavirus A | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Jayawardena N / Burga L | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2018 Journal: Proc Natl Acad Sci U S A / Year: 2018Title: Structural basis for anthrax toxin receptor 1 recognition by Seneca Valley Virus. Authors: Nadishka Jayawardena / Laura N Burga / Richard A Easingwood / Yoshimasa Takizawa / Matthias Wolf / Mihnea Bostina /   Abstract: Recently, the use of oncolytic viruses in cancer therapy has become a realistic therapeutic option. Seneca Valley Virus (SVV) is a newly discovered picornavirus, which has earned a significant ...Recently, the use of oncolytic viruses in cancer therapy has become a realistic therapeutic option. Seneca Valley Virus (SVV) is a newly discovered picornavirus, which has earned a significant reputation as a potent oncolytic agent. Anthrax toxin receptor 1 (ANTXR1), one of the cellular receptors for the protective antigen secreted by , has been identified as the high-affinity cellular receptor for SVV. Here, we report the structure of the SVV-ANTXR1 complex determined by single-particle cryo-electron microscopy analysis at near-atomic resolution. This is an example of a shared receptor structure between a mammalian virus and a bacterial toxin. Our structure shows that ANTXR1 decorates the outer surface of the SVV capsid and interacts with the surface-exposed BC loop and loop II of VP1, "the puff" of VP2 and "the knob" of VP3. Comparison of the receptor-bound capsid structure with the native capsid structure reveals that receptor binding induces minor conformational changes in SVV capsid structure, suggesting the role of ANTXR1 as an attachment receptor. Furthermore, our results demonstrate that the capsid footprint on the receptor is not conserved in anthrax toxin receptor 2 (ANTXR2), thereby providing a molecular mechanism for explaining the exquisite selectivity of SVV for ANTXR1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7772.map.gz emd_7772.map.gz | 64.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7772-v30.xml emd-7772-v30.xml emd-7772.xml emd-7772.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7772.png emd_7772.png | 244.1 KB | ||
| Filedesc metadata |  emd-7772.cif.gz emd-7772.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7772 http://ftp.pdbj.org/pub/emdb/structures/EMD-7772 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7772 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7772 | HTTPS FTP |
-Related structure data
| Related structure data |  6cx1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7772.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7772.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Seneca Valley Virus-Anthrax Toxin Receptor 1 complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.42 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Seneca Valley Virus-Anthrax Toxin Receptor 1 complex
| Entire | Name: Seneca Valley Virus-Anthrax Toxin Receptor 1 complex |
|---|---|
| Components |
|
-Supramolecule #1: Senecavirus A
| Supramolecule | Name: Senecavirus A / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 390157 / Sci species name: Senecavirus A / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Molecular weight | Theoretical: 6.5 MDa |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 300.0 Å / T number (triangulation number): 3 |
-Macromolecule #1: Anthrax toxin receptor 1
| Macromolecule | Name: Anthrax toxin receptor 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 21.026668 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: CYGGFDLYFI LDKSGSVLHH WNEIYYFVEQ LAHKFISPQL RMSFIVFSTR GTTLMKLTED REQIRQGLEE LQKVLPGGDT YMHEGFERA SEQIYYENRQ GYRTASVIIA LTDGELHEDL FFYSEREANR SRDLGAIVYA VGVKDFNETQ LARIADSKDH V FPVNDGFQ ALQGIIHSIL KKSC UniProtKB: Anthrax toxin receptor 1 |
-Macromolecule #2: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Senecavirus A Senecavirus A |
| Molecular weight | Theoretical: 28.459969 KDa |
| Sequence | String: STDNAETGVI EAGNTDTDFS GELAAPGSNH TNVKFLFDRS RLLNVIKVLE KDAVFPRPFP TQEGAQQDDG YFCLLTPRPT VASRPATRF GLYANPSGSG VLANTSLDFN FYSLACFTYF RSDLEVTVVS LEPDLEFAVG WFPSGSEYQA SSFVYDQLHV P FHFTGRTP ...String: STDNAETGVI EAGNTDTDFS GELAAPGSNH TNVKFLFDRS RLLNVIKVLE KDAVFPRPFP TQEGAQQDDG YFCLLTPRPT VASRPATRF GLYANPSGSG VLANTSLDFN FYSLACFTYF RSDLEVTVVS LEPDLEFAVG WFPSGSEYQA SSFVYDQLHV P FHFTGRTP RAFASKGGKV SFVLPWNSVS SVLPVRWGGA SKLSSATRGL PAHADWGTIY AFVPRPNEKK STAVKHVAVY IR YKNARAW CPSMLPFRSY K UniProtKB: Genome polyprotein |
-Macromolecule #3: Capsid protein VP2
| Macromolecule | Name: Capsid protein VP2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Senecavirus A Senecavirus A |
| Molecular weight | Theoretical: 29.870316 KDa |
| Sequence | String: DRVTTQTAGN TAINTQSSLG VLCAYVEDPT KSDPPSSSTD QPTTTFTAID RWYTGRLNSW TKAVKTFSFQ AVPLPGAFLS RQGGLNGGA FTATLHRHFL MKCGWQVQVQ CNLTQFHQGA LLVAMVPETT LDVKPDGKAK SLQELNEEQW VEMSDDYRTG K NMPFQSLG ...String: DRVTTQTAGN TAINTQSSLG VLCAYVEDPT KSDPPSSSTD QPTTTFTAID RWYTGRLNSW TKAVKTFSFQ AVPLPGAFLS RQGGLNGGA FTATLHRHFL MKCGWQVQVQ CNLTQFHQGA LLVAMVPETT LDVKPDGKAK SLQELNEEQW VEMSDDYRTG K NMPFQSLG TYYRPPNWTW GPNFINPYQV TVFPHQILNA RTSTSVDINV PYIGETPTQS SETQNSWTLL VMVLVPLDYK EG ATTDPEI TFSVRPTSPY FNGLRNRYTA G UniProtKB: Genome polyprotein |
-Macromolecule #4: Capsid protein VP3
| Macromolecule | Name: Capsid protein VP3 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Senecavirus A Senecavirus A |
| Molecular weight | Theoretical: 26.353938 KDa |
| Sequence | String: GPIPTAPREN SLMFLSTLPD DTVPAYGNVR TPPVNYLPGE ITDLLQLARI PTLMAFERVP EPVPASDTYV PYVAVPTQFD DRPLISFPI TLSDPVYQNT LVGAISSNFA NYRGCIQITL TFCGPMMARG KFLLSYSPPN GTQPQTLSEA MQCTYSIWDI G LNSSWTFV ...String: GPIPTAPREN SLMFLSTLPD DTVPAYGNVR TPPVNYLPGE ITDLLQLARI PTLMAFERVP EPVPASDTYV PYVAVPTQFD DRPLISFPI TLSDPVYQNT LVGAISSNFA NYRGCIQITL TFCGPMMARG KFLLSYSPPN GTQPQTLSEA MQCTYSIWDI G LNSSWTFV VPYISPSDYR ETRAITNSVY SADGWFSLHK LTKITLPPDC PQSPCILFFA SAGEDYTLRL PVDCNPSYVF UniProtKB: Genome polyprotein |
-Macromolecule #5: Capsid protein VP4
| Macromolecule | Name: Capsid protein VP4 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Senecavirus A Senecavirus A |
| Molecular weight | Theoretical: 5.998467 KDa |
| Sequence | String: RGNNGNMTFN YYANTYQNSV DFSTSSSASG AGPGNSRGGL AGLLTNFSGI LNPLGYLK UniProtKB: Genome polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Name: Phosphate Buffer |
| Grid | Model: Quantifoil R1.2/1.3 / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 12 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV / Details: Blot force 0, 3 sec blotting time. |
| Details | A homogenous sample containing full capsids |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 39.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 73000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 100 / Target criteria: CC |
|---|---|
| Output model |  PDB-6cx1: |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















