+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12154 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Kashmir bee virus empty particle at acidic pH | |||||||||
 Map data Map data | KBV empty particle at acidic pH | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Viruses / Riboviria / Orthornavirae / Pisuviricota / Pisoniviricetes / Picornavirales / Dicistroviridae / Aparavirus / Kashmir Bee Virus / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Kashmir bee virus Kashmir bee virus | |||||||||
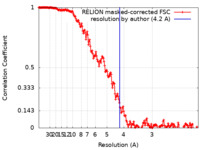
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Mukhamedova L / Plevka P | |||||||||
| Funding support |  Czech Republic, 2 items Czech Republic, 2 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2021 Journal: J Virol / Year: 2021Title: Virion structure and genome release mechanism of dicistrovirus Kashmir bee virus. Authors: Liya Mukhamedova / Tibor Füzik / Jiří Nováček / Dominik Hrebík / Antonín Přidal / Gerardo A Marti / Diego M A Guérin / Pavel Plevka /    Abstract: Infections of Kashmir bee virus (KBV) are lethal for honeybees and have been associated with colony collapse disorder. KBV and closely related viruses contribute to the ongoing decline in the number ...Infections of Kashmir bee virus (KBV) are lethal for honeybees and have been associated with colony collapse disorder. KBV and closely related viruses contribute to the ongoing decline in the number of honeybee colonies in North America, Europe, Australia, and other parts of the world. Despite the economic and ecological impact of KBV, its structure and infection process remain unknown. Here we present the structure of the virion of KBV determined to a resolution of 2.8 Å. We show that the exposure of KBV to acidic pH induces a reduction in inter-pentamer contacts within capsids and the reorganization of its RNA genome from a uniform distribution to regions of high and low density. Capsids of KBV crack into pieces at acidic pH, resulting in the formation of open particles lacking pentamers of capsid proteins. The large openings of capsids enable the rapid release of genomes and thus limit the probability of their degradation by RNases. The opening of capsids may be a shared mechanism for the genome release of viruses from the family The western honeybee () is indispensable for maintaining agricultural productivity as well as the abundance and diversity of wild flowering plants. However, bees suffer from environmental pollution, parasites, and pathogens, including viruses. Outbreaks of virus infections cause the deaths of individual honeybees as well as collapses of whole colonies. Kashmir bee virus has been associated with colony collapse disorder in the US, and no cure of the disease is currently available. Here we report the structure of an infectious particle of Kashmir bee virus and show how its protein capsid opens to release the genome. Our structural characterization of the infection process determined that therapeutic compounds stabilizing contacts between pentamers of capsid proteins could prevent the genome release of the virus. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12154.map.gz emd_12154.map.gz | 59 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12154-v30.xml emd-12154-v30.xml emd-12154.xml emd-12154.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12154_fsc.xml emd_12154_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_12154.png emd_12154.png | 278.6 KB | ||
| Filedesc metadata |  emd-12154.cif.gz emd-12154.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12154 http://ftp.pdbj.org/pub/emdb/structures/EMD-12154 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12154 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12154 | HTTPS FTP |
-Related structure data
| Related structure data |  7be9MC  7bc3C  7bg8C  7bgkC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12154.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12154.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | KBV empty particle at acidic pH | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Kashmir bee virus
| Entire | Name:  Kashmir bee virus Kashmir bee virus |
|---|---|
| Components |
|
-Supramolecule #1: Kashmir bee virus
| Supramolecule | Name: Kashmir bee virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 68876 / Sci species name: Kashmir bee virus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Molecular weight | Theoretical: 5.932 MDa |
| Virus shell | Shell ID: 1 / Name: Full virus / Diameter: 350.0 Å / T number (triangulation number): 3 |
-Macromolecule #1: Structural polyprotein
| Macromolecule | Name: Structural polyprotein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Kashmir bee virus Kashmir bee virus |
| Molecular weight | Theoretical: 22.735377 KDa |
| Sequence | String: ENTISFFDSG DPERMNNEAL MRGCGEQIVN LRPLLRTFRT INDNWSLAAN TKTPITDLTN TADAEGRDYM SYLSFLYRFY RGGRRYKFF NTTPLKQSQT CYVRSFLIPR NYTADEINTD GPSHITYPVI NPVHEVEVPF YSQYRKIPIA STSDKGYDSS L MYYTNVGT ...String: ENTISFFDSG DPERMNNEAL MRGCGEQIVN LRPLLRTFRT INDNWSLAAN TKTPITDLTN TADAEGRDYM SYLSFLYRFY RGGRRYKFF NTTPLKQSQT CYVRSFLIPR NYTADEINTD GPSHITYPVI NPVHEVEVPF YSQYRKIPIA STSDKGYDSS L MYYTNVGT QQIVARAGND DFTFGWMIGT PQLQGITKEV UniProtKB: Structural polyprotein |
-Macromolecule #2: Structural polyprotein
| Macromolecule | Name: Structural polyprotein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Kashmir bee virus Kashmir bee virus |
| Molecular weight | Theoretical: 31.920127 KDa |
| Sequence | String: SKPRNQNQVM PYQNVPGWGY SLYKGIDMSV PLAYDPNNEL GDLRDVFPSA VDEMAIGYVC GNPAIKHVLT WSTTDVVQNP ISNGDDWGG VIPVGMPCYS KTIRAVKGAT STSKTEVMDP APCEYVANLF SYWRATMCYR ITVVKTAFHT GRLEIFFEPG S IPTVRTAD ...String: SKPRNQNQVM PYQNVPGWGY SLYKGIDMSV PLAYDPNNEL GDLRDVFPSA VDEMAIGYVC GNPAIKHVLT WSTTDVVQNP ISNGDDWGG VIPVGMPCYS KTIRAVKGAT STSKTEVMDP APCEYVANLF SYWRATMCYR ITVVKTAFHT GRLEIFFEPG S IPTVRTAD NLGPDQTQLN GTIAPSDNNY KYILDLTNDT EVTIKVPYVS NKMFMKTVGI YGAHDEDNWN FDESFTGFLC IR PITKLMA PDTVSQKVSI VVWKWAEDVV VVEPKPLTSG PTQVYNPPAV A UniProtKB: Structural polyprotein |
-Macromolecule #3: Structural polyprotein
| Macromolecule | Name: Structural polyprotein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Kashmir bee virus Kashmir bee virus |
| Molecular weight | Theoretical: 21.998068 KDa |
| Sequence | String: SIIQFLQRPV LIDNIEIVAG TTADNNTALS RYVLDRTNPQ KYIKQWTLPS TVLKAGGKAQ KLANFKYLRC DVQVKIVLNA NPFIAGRLY LAYSPYDDKV APERRIIYTS RAGVTGYPGV ELDFQLDNSV EMTIPYASFQ EAYDLVSGNE DFVQLYLFTI A PVLGPSAE ...String: SIIQFLQRPV LIDNIEIVAG TTADNNTALS RYVLDRTNPQ KYIKQWTLPS TVLKAGGKAQ KLANFKYLRC DVQVKIVLNA NPFIAGRLY LAYSPYDDKV APERRIIYTS RAGVTGYPGV ELDFQLDNSV EMTIPYASFQ EAYDLVSGNE DFVQLYLFTI A PVLGPSAE SANSKVDLSV YMWLDNISLV IPTYRLNP UniProtKB: Structural polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | tissue |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 6 |
| Grid | Model: Quantifoil / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 103.15 K / Max: 108.15 K |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 8193 / Average exposure time: 1.0 sec. / Average electron dose: 94.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 4.134 µm / Calibrated defocus min: 0.214 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.2 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)