+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3715 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | T=4 HBV virus-like particle. | |||||||||
 Map data Map data | T=4 Hepatitis B virus virus-like particle | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Hepatitis B virus Hepatitis B virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Patel N / White SJ / Thompson RF / Bingham R / Weiss EU / Maskell DP / Zlotnick A / Dykeman E / Tuma R / Twarock R ...Patel N / White SJ / Thompson RF / Bingham R / Weiss EU / Maskell DP / Zlotnick A / Dykeman E / Tuma R / Twarock R / Ranson NA / Stockley PG | |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2017 Journal: Nat Microbiol / Year: 2017Title: HBV RNA pre-genome encodes specific motifs that mediate interactions with the viral core protein that promote nucleocapsid assembly. Authors: Nikesh Patel / Simon J White / Rebecca F Thompson / Richard Bingham / Eva U Weiß / Daniel P Maskell / Adam Zlotnick / Eric Dykeman / Roman Tuma / Reidun Twarock / Neil A Ranson / Peter G Stockley /   Abstract: Formation of the hepatitis B virus nucleocapsid is an essential step in the viral lifecycle, but its assembly is not fully understood. We report the discovery of sequence-specific interactions ...Formation of the hepatitis B virus nucleocapsid is an essential step in the viral lifecycle, but its assembly is not fully understood. We report the discovery of sequence-specific interactions between the viral pre-genome and the hepatitis B core protein that play roles in defining the nucleocapsid assembly pathway. Using RNA SELEX and bioinformatics, we identified multiple regions in the pre-genomic RNA with high affinity for core protein dimers. These RNAs form stem-loops with a conserved loop motif that trigger sequence-specific assembly of virus-like particles (VLPs) at much higher fidelity and yield than in the absence of RNA. The RNA oligos do not interact with preformed RNA-free VLPs, so their effects must occur during particle assembly. Asymmetric cryo-electron microscopy reconstruction of the T = 4 VLPs assembled in the presence of one of the RNAs reveals a unique internal feature connected to the main core protein shell via lobes of density. Biophysical assays suggest that this is a complex involving several RNA oligos interacting with the C-terminal arginine-rich domains of core protein. These core protein-RNA contacts may play one or more roles in regulating the organization of the pre-genome during nucleocapsid assembly, facilitating subsequent reverse transcription and acting as a nucleation complex for nucleocapsid assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3715.map.gz emd_3715.map.gz | 78.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3715-v30.xml emd-3715-v30.xml emd-3715.xml emd-3715.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3715_fsc.xml emd_3715_fsc.xml | 13.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_3715.png emd_3715.png | 204.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3715 http://ftp.pdbj.org/pub/emdb/structures/EMD-3715 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3715 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3715 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3715.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3715.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | T=4 Hepatitis B virus virus-like particle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
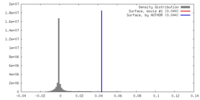
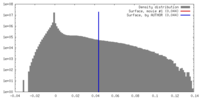
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Hepatitis B virus
| Entire | Name:   Hepatitis B virus Hepatitis B virus |
|---|---|
| Components |
|
-Supramolecule #1: Hepatitis B virus
| Supramolecule | Name: Hepatitis B virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Is adw strain but with some mutations from deposited sequence. NCBI-ID: 10407 / Sci species name: Hepatitis B virus / Sci species strain: adw / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host system | Organism:  |
| Virus shell | Shell ID: 1 / T number (triangulation number): 4 |
-Macromolecule #1: Hepatitis B virus core antigen
| Macromolecule | Name: Hepatitis B virus core antigen / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Hepatitis B virus / Strain: adw Hepatitis B virus / Strain: adw |
| Recombinant expression | Organism:  |
| Sequence | String: MDIDPYKEFG ATVELLSFL P SDFFPSVR DL LDTASAL YRE ALESPE HCSP HHTAL RQAIL CWGE LMTLAT WVG NNLEDPA SR DLVVNYVN T NMGLKIRQL LWFHISCLTF GRETVLEYL V SFGVWIRT PP AYRPPNA PIL STLPET TVVR RRDRG ...String: MDIDPYKEFG ATVELLSFL P SDFFPSVR DL LDTASAL YRE ALESPE HCSP HHTAL RQAIL CWGE LMTLAT WVG NNLEDPA SR DLVVNYVN T NMGLKIRQL LWFHISCLTF GRETVLEYL V SFGVWIRT PP AYRPPNA PIL STLPET TVVR RRDRG RSPRR RTPS PRRRRS QSP RRRRSQS RE SQC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.2 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 5.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 281 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 2397 / Average exposure time: 2.5 sec. / Average electron dose: 67.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)