[English] 日本語
 Yorodumi
Yorodumi- EMDB-6710: Structure of the 30S small subunit of chloroplast ribosome from s... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6710 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the 30S small subunit of chloroplast ribosome from spinach | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cryo-EM / ribosome / chloroplast ribosome | |||||||||
| Function / homology |  Function and homology information Function and homology informationchloroplast rRNA processing / plastid small ribosomal subunit / plastid translation / negative regulation of translational elongation / mitochondrial small ribosomal subunit / mitochondrial translation / chloroplast stroma / plastid / chloroplast thylakoid membrane / ribosomal small subunit binding ...chloroplast rRNA processing / plastid small ribosomal subunit / plastid translation / negative regulation of translational elongation / mitochondrial small ribosomal subunit / mitochondrial translation / chloroplast stroma / plastid / chloroplast thylakoid membrane / ribosomal small subunit binding / chloroplast / ribosomal small subunit biogenesis / ribosomal small subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / response to antibiotic / mRNA binding / mitochondrion / RNA binding Similarity search - Function | |||||||||
| Biological species |  Spinacia oleracea (spinach) Spinacia oleracea (spinach) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Ahmed T / Shi J | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2017 Journal: Nucleic Acids Res / Year: 2017Title: Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation. Authors: Tofayel Ahmed / Jian Shi / Shashi Bhushan /  Abstract: Chloroplastic translation is mediated by a bacterial-type 70S chloroplast ribosome. During the evolution, chloroplast ribosomes have acquired five plastid-specific ribosomal proteins or PSRPs (cS22, ...Chloroplastic translation is mediated by a bacterial-type 70S chloroplast ribosome. During the evolution, chloroplast ribosomes have acquired five plastid-specific ribosomal proteins or PSRPs (cS22, cS23, bTHXc, cL37 and cL38) which have been suggested to play important regulatory roles in translation. However, their exact locations on the chloroplast ribosome remain elusive due to lack of a high-resolution structure, hindering our progress to understand their possible roles. Here we present a cryo-EM structure of the 70S chloroplast ribosome from spinach resolved to 3.4 Å and focus our discussion mainly on the architecture of the 30S small subunit (SSU) which is resolved to 3.7 Å. cS22 localizes at the SSU foot where it seems to compensate for the deletions in 16S rRNA. The mRNA exit site is highly remodeled due to the presence of cS23 suggesting an alternative mode of translation initiation. bTHXc is positioned at the SSU head and appears to stabilize the intersubunit bridge B1b during thermal fluctuations. The translation factor plastid pY binds to the SSU on the intersubunit side and interacts with the conserved nucleotide bases involved in decoding. Most of the intersubunit bridges are conserved compared to the bacteria, except for a new bridge involving uL2c and bS6c. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6710.map.gz emd_6710.map.gz | 9.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6710-v30.xml emd-6710-v30.xml emd-6710.xml emd-6710.xml | 38.2 KB 38.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6710.png emd_6710.png | 81 KB | ||
| Filedesc metadata |  emd-6710.cif.gz emd-6710.cif.gz | 9.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6710 http://ftp.pdbj.org/pub/emdb/structures/EMD-6710 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6710 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6710 | HTTPS FTP |
-Validation report
| Summary document |  emd_6710_validation.pdf.gz emd_6710_validation.pdf.gz | 389.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6710_full_validation.pdf.gz emd_6710_full_validation.pdf.gz | 389 KB | Display | |
| Data in XML |  emd_6710_validation.xml.gz emd_6710_validation.xml.gz | 7 KB | Display | |
| Data in CIF |  emd_6710_validation.cif.gz emd_6710_validation.cif.gz | 8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6710 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6710 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6710 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6710 | HTTPS FTP |
-Related structure data
| Related structure data |  5x8rMC  6709C  6711C  5x8pC  5x8tC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6710.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6710.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
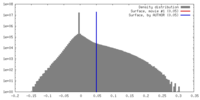
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : 30S small subunit of chloroplast ribosome from spinach
+Supramolecule #1: 30S small subunit of chloroplast ribosome from spinach
+Macromolecule #1: 30S ribosomal protein S2, chloroplastic
+Macromolecule #2: 30S ribosomal protein S3, chloroplastic
+Macromolecule #3: 30S ribosomal protein S5, chloroplastic
+Macromolecule #4: 30S ribosomal protein S6 alpha, chloroplastic
+Macromolecule #5: 30S ribosomal protein S7, chloroplastic
+Macromolecule #6: 30S ribosomal protein S8, chloroplastic
+Macromolecule #7: 30S ribosomal protein S9, chloroplastic
+Macromolecule #8: protein S10
+Macromolecule #9: 30S ribosomal protein S11, chloroplastic
+Macromolecule #10: 30S ribosomal protein S12, chloroplastic
+Macromolecule #11: 30S ribosomal protein S13, chloroplastic
+Macromolecule #12: 30S ribosomal protein S15, chloroplastic
+Macromolecule #13: 30S ribosomal protein S16, chloroplastic
+Macromolecule #14: protein S17
+Macromolecule #15: 30S ribosomal protein S18, chloroplastic
+Macromolecule #16: 30S ribosomal protein S19 alpha, chloroplastic
+Macromolecule #17: protein S20
+Macromolecule #18: protein S21
+Macromolecule #19: protein plastid pY
+Macromolecule #21: protein cS23
+Macromolecule #22: 30S ribosomal protein S4, chloroplastic
+Macromolecule #23: protein cS22
+Macromolecule #24: 30S ribosomal protein S14, chloroplastic
+Macromolecule #25: protein bTHXc
+Macromolecule #26: 30S ribosomal protein S1, chloroplastic
+Macromolecule #20: 16S rRNA
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 Details: 20 mM Tris HCl, pH 7.6, 100 mM KCl, 10 mM MgOAc, 100 mM sucrose, 7 mM 2-mercaptoethanol, 1 unit/ml RNase inhibitor, 0.1% protease inhibitor |
|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Frames/image: 1-25 / Number grids imaged: 1 / Number real images: 3161 / Average electron dose: 1.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 3.7 µm / Calibrated defocus min: 0.4 µm / Calibrated magnification: 133333 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.7 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)