+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5557 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the EV71 strain 1095 procapsid | |||||||||
 Map data Map data | Reconstruction of EV71 strain 1095 procapsid | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | picornavirus / enterovirus 71 / hand foot and mouth disease / cryo-EM / single particle reconstruction | |||||||||
| Biological species |   Human enterovirus 71 Human enterovirus 71 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.78 Å | |||||||||
 Authors Authors | Cifuente JO / Lee H / Yoder JD / Shingler KL / Carnegie MS / Yoder JL / Ashley RE / Makhov AM / Conway JF / Hafenstein S | |||||||||
 Citation Citation |  Journal: J Virol / Year: 2013 Journal: J Virol / Year: 2013Title: Structures of the procapsid and mature virion of enterovirus 71 strain 1095. Authors: Javier O Cifuente / Hyunwook Lee / Joshua D Yoder / Kristin L Shingler / Michael S Carnegie / Jennifer L Yoder / Robert E Ashley / Alexander M Makhov / James F Conway / Susan Hafenstein /  Abstract: Enterovirus 71 (EV71) is an important emerging human pathogen with a global distribution and presents a disease pattern resembling poliomyelitis with seasonal epidemics that include cases of severe ...Enterovirus 71 (EV71) is an important emerging human pathogen with a global distribution and presents a disease pattern resembling poliomyelitis with seasonal epidemics that include cases of severe neurological complications, such as acute flaccid paralysis. EV71 is a member of the Picornaviridae family, which consists of icosahedral, nonenveloped, single-stranded RNA viruses. Here we report structures derived from X-ray crystallography and cryoelectron microscopy (cryo-EM) for the 1095 strain of EV71, including a putative precursor in virus assembly, the procapsid, and the mature virus capsid. The cryo-EM map of the procapsid provides new structural information on portions of the capsid proteins VP0 and VP1 that are disordered in the higher-resolution crystal structures. Our structures solved from virus particles in solution are largely in agreement with those from prior X-ray crystallographic studies; however, we observe small but significant structural differences for the 1095 procapsid compared to a structure solved in a previous study (X. Wang, W. Peng, J. Ren, Z. Hu, J. Xu, Z. Lou, X. Li, W. Yin, X. Shen, C. Porta, T. S. Walter, G. Evans, D. Axford, R. Owen, D. J. Rowlands, J. Wang, D. I. Stuart, E. E. Fry, and Z. Rao, Nat. Struct. Mol. Biol. 19:424-429, 2012) for a different strain of EV71. For both EV71 strains, the procapsid is significantly larger in diameter than the mature capsid, unlike in any other picornavirus. Nonetheless, our results demonstrate that picornavirus capsid expansion is possible without RNA encapsidation and that picornavirus assembly may involve an inward radial collapse of the procapsid to yield the native virion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5557.map.gz emd_5557.map.gz | 36.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5557-v30.xml emd-5557-v30.xml emd-5557.xml emd-5557.xml | 11.3 KB 11.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5557_1.jpg emd_5557_1.jpg | 127.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5557 http://ftp.pdbj.org/pub/emdb/structures/EMD-5557 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5557 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5557 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5557.map.gz / Format: CCP4 / Size: 154.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5557.map.gz / Format: CCP4 / Size: 154.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of EV71 strain 1095 procapsid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2545 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
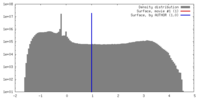
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : EV71 strain 1095 procapsid
| Entire | Name: EV71 strain 1095 procapsid |
|---|---|
| Components |
|
-Supramolecule #1000: EV71 strain 1095 procapsid
| Supramolecule | Name: EV71 strain 1095 procapsid / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Human enterovirus 71
| Supramolecule | Name: Human enterovirus 71 / type: virus / ID: 1 / NCBI-ID: 39054 / Sci species name: Human enterovirus 71 / Database: NCBI / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
| Molecular weight | Theoretical: 8 MDa |
| Virus shell | Shell ID: 1 / Diameter: 300 Å / T number (triangulation number): 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: 10mM Tris-HCl, 200mM NaCl, 50mM MgCl2 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE MIXTURE / Chamber temperature: 120 K / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 83 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 100,000 times magnification |
| Specialist optics | Energy filter - Name: FEI |
| Date | Aug 15, 2011 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 6.35 µm / Number real images: 20 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 50400 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 3.91 µm / Nominal defocus min: 0.79 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Semi-automatic particle selection was performed using e2boxer.py to obtain the particle coordinates, followed by particle boxing, linearization, normalization, and apodization of the images using Robem. Defocus and astigmatism values to perform contrast transfer function (CTF) correction were assessed using Robem for the extracted particles. The icosahedrally averaged reconstructions were initiated using a random model generated with setup_rmc and reached better than 10 A resolution estimated at a Fourier Shell Correlation, FSC=0.5. For the last step of refinement, the final maps were CTF corrected using a B factor of 300 A2. |
|---|---|
| CTF correction | Details: Each particle |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 8.78 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Auto3dem, EMAN2 / Number images used: 8805 |
| Final angle assignment | Details: AUTO3DEM |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B / Chain - #2 - Chain ID: C |
|---|---|
| Software | Name: VEDA |
| Details | Protocol: Rigid body |
| Refinement | Space: RECIPROCAL / Protocol: RIGID BODY FIT / Target criteria: R-factor |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)