+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3574 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of deformed wing virus, a honeybee pathogen | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Deformed wing virus / Picornavirales / Iflaviridae / Iflavirus / viral protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral capsid / RNA helicase activity / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / structural molecule activity / proteolysis / RNA binding / ATP binding Similarity search - Function | |||||||||
| Biological species |   Deformed wing virus Deformed wing virus | |||||||||
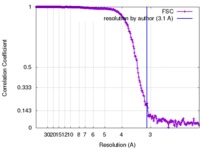
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Skubnik K / Novacek J | |||||||||
| Funding support | 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2017 Journal: Proc Natl Acad Sci U S A / Year: 2017Title: Structure of deformed wing virus, a major honey bee pathogen. Authors: Karel Škubník / Jiří Nováček / Tibor Füzik / Antonín Přidal / Robert J Paxton / Pavel Plevka /   Abstract: The worldwide population of western honey bees () is under pressure from habitat loss, environmental stress, and pathogens, particularly viruses that cause lethal epidemics. Deformed wing virus (DWV) ...The worldwide population of western honey bees () is under pressure from habitat loss, environmental stress, and pathogens, particularly viruses that cause lethal epidemics. Deformed wing virus (DWV) from the family , together with its vector, the mite , is likely the major threat to the world's honey bees. However, lack of knowledge of the atomic structures of iflaviruses has hindered the development of effective treatments against them. Here, we present the virion structures of DWV determined to a resolution of 3.1 Å using cryo-electron microscopy and 3.8 Å by X-ray crystallography. The C-terminal extension of capsid protein VP3 folds into a globular protruding (P) domain, exposed on the virion surface. The P domain contains an Asp-His-Ser catalytic triad that is, together with five residues that are spatially close, conserved among iflaviruses. These residues may participate in receptor binding or provide the protease, lipase, or esterase activity required for entry of the virus into a host cell. Furthermore, nucleotides of the DWV RNA genome interact with VP3 subunits. The capsid protein residues involved in the RNA binding are conserved among honey bee iflaviruses, suggesting a putative role of the genome in stabilizing the virion or facilitating capsid assembly. Identifying the RNA-binding and putative catalytic sites within the DWV virion structure enables future analyses of how DWV and other iflaviruses infect insect cells and also opens up possibilities for the development of antiviral treatments. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3574.map.gz emd_3574.map.gz | 59 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3574-v30.xml emd-3574-v30.xml emd-3574.xml emd-3574.xml | 23.4 KB 23.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3574_fsc.xml emd_3574_fsc.xml | 22.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_3574.png emd_3574.png | 228.1 KB | ||
| Filedesc metadata |  emd-3574.cif.gz emd-3574.cif.gz | 7.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3574 http://ftp.pdbj.org/pub/emdb/structures/EMD-3574 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3574 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3574 | HTTPS FTP |
-Related structure data
| Related structure data |  5mv5MC  3570C  3575C  4009C  4014C  5g51C  5g52C  5l7qC  5l8qC  5mupC  5mv6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3574.map.gz / Format: CCP4 / Size: 620.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3574.map.gz / Format: CCP4 / Size: 620.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.054 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Deformed wing virus
| Entire | Name:   Deformed wing virus Deformed wing virus |
|---|---|
| Components |
|
-Supramolecule #1: Deformed wing virus
| Supramolecule | Name: Deformed wing virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 / Details: Virus was purified from honeybee pupae. / NCBI-ID: 198112 / Sci species name: Deformed wing virus / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / Diameter: 390.0 Å / T number (triangulation number): 3 |
-Macromolecule #1: VP1
| Macromolecule | Name: VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Deformed wing virus Deformed wing virus |
| Molecular weight | Theoretical: 28.679273 KDa |
| Sequence | String: GEESRNTTVL DTTTTLQSSG FGRAFFGEAF NDLKTLMRRY QLYGQLLLSV TTDKDIDHCM FTFPCLPQGL ALDIGSAGSP HEIFNRCRD GIIPLIASGY RFYRGDLRYK IVFPSNVNSN IWVQHRPDRR LEGWSAAKIV NCDAVSTGQG VYNHGYASHI Q ITRVNNVI ...String: GEESRNTTVL DTTTTLQSSG FGRAFFGEAF NDLKTLMRRY QLYGQLLLSV TTDKDIDHCM FTFPCLPQGL ALDIGSAGSP HEIFNRCRD GIIPLIASGY RFYRGDLRYK IVFPSNVNSN IWVQHRPDRR LEGWSAAKIV NCDAVSTGQG VYNHGYASHI Q ITRVNNVI ELEVPFYNAT CYNYLQAFNA SSAASSYAVS LGEISVGFQA TSDDIASIVN KPVTIYYSIG DGMQFSQWVG YQ PMMILDQ LPAPVVRAVP E UniProtKB: Genome polyprotein |
-Macromolecule #2: VP2
| Macromolecule | Name: VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Deformed wing virus Deformed wing virus |
| Molecular weight | Theoretical: 28.3609 KDa |
| Sequence | String: MDNPNPGPDG EGEVELEKDS NVVLTTQRDP STSIPAPVSV KWSRWTSNDV VDDYATITSR WYQIAEFVWS KDDPFDKELA RLILPRALL SSIEANSDAI CDVPNTIPFK VHAYWRGDME VRVQINSNKF QVGQLQATWY YSDHENLNIS SKRSVYGFSQ M DHALISAS ...String: MDNPNPGPDG EGEVELEKDS NVVLTTQRDP STSIPAPVSV KWSRWTSNDV VDDYATITSR WYQIAEFVWS KDDPFDKELA RLILPRALL SSIEANSDAI CDVPNTIPFK VHAYWRGDME VRVQINSNKF QVGQLQATWY YSDHENLNIS SKRSVYGFSQ M DHALISAS ASNEAKLVIP FKHVYPFLPT RIVPDWTTGI LDMGALNIRV IAPLRMSATG PTTCNVVVFI KLNNSEFTGT SS GKFYASQ IRAKPE UniProtKB: Genome polyprotein |
-Macromolecule #3: VP3
| Macromolecule | Name: VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Deformed wing virus Deformed wing virus |
| Molecular weight | Theoretical: 46.697582 KDa |
| Sequence | String: DNPSYQQSPR HFVPTGMHSL ALGTNLVEPL HALRLDAAGT TQHPVGCAPD EDMTVSSIAS RYGLIRRVQW KKDHAKGSLL LQLDADPFV EQRIEGTNPI SLYWFAPVGV VSSMFMQWRG SLEYRFDIIA SQFHTGRLIV GYVPGLTASL QLQMDYMKLK S SSYVVFDL ...String: DNPSYQQSPR HFVPTGMHSL ALGTNLVEPL HALRLDAAGT TQHPVGCAPD EDMTVSSIAS RYGLIRRVQW KKDHAKGSLL LQLDADPFV EQRIEGTNPI SLYWFAPVGV VSSMFMQWRG SLEYRFDIIA SQFHTGRLIV GYVPGLTASL QLQMDYMKLK S SSYVVFDL QESNSFTFEV PYVSYRPWWV RKYGGNYLPS STDAPSTLFM YVQVPLIPME AVSDTIDINV YVRGGSSFEV CV PVQPSLG LNWNTDFILR NDEEYRAKTG YAPYYAGVWH SFNNSNSLVF RWGSASDQIA QWPTISVPRG ELAFLRIKDG KQA AVGTQP WRTMVVWPSG HGYNIGIPTY NAERARQLAQ HLYGGGSLTD EKAKQLFVPA NQQGPGKVSN GNPVWEVMRA PLAT QRAHI QDFEFIEAIP E UniProtKB: Genome polyprotein |
-Macromolecule #4: URIDINE-5'-MONOPHOSPHATE
| Macromolecule | Name: URIDINE-5'-MONOPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: U5P |
|---|---|
| Molecular weight | Theoretical: 324.181 Da |
| Chemical component information |  ChemComp-U: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: Dulbeccos Phosphate Buffered Saline D8537 sigma aldrich |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: NITROGEN / Pretreatment - Pressure: 0.007 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
| Details | Virus was incubated in high salt solution containing 0.8 M potassium dihydrogen phosphate, 0.8 M sodium dihydrogen phosphate, 0.1 M sodium HEPES, pH 7.5. After 12 hours incubation glutaraldehyde was added to final concentration 1%. After 1 hour was virus dialysed into PBS buffer. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Frames/image: 2-16 / Number grids imaged: 1 / Average exposure time: 1.0 sec. / Average electron dose: 21.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 74235 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER / Target criteria: R-factor |
|---|---|
| Output model |  PDB-5mv5: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)