[English] 日本語
 Yorodumi
Yorodumi- EMDB-52559: Mycobacterium smegmatis inosine monophosphate dehydrogenase (IMPD... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mycobacterium smegmatis inosine monophosphate dehydrogenase (IMPDH) E-XMP* intermediate, compressed | |||||||||
 Map data Map data | LocScale primary viewing map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Octamer / reaction intermediate / Purine metabolism / IMPDH / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationIMP dehydrogenase / IMP dehydrogenase activity / GMP biosynthetic process / GTP biosynthetic process / nucleotide binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.73 Å | |||||||||
 Authors Authors | Bulvas O / Kouba T / Pichova I | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2025 Journal: J Struct Biol / Year: 2025Title: Conformational landscape of the mycobacterial inosine 5'-monophosphate dehydrogenase octamerization interface. Authors: Ondřej Bulvas / Zdeněk Knejzlík / Anatolij Filimoněnko / Tomáš Kouba / Iva Pichová /  Abstract: Inosine 5'-monophosphate dehydrogenase (IMPDH), a key enzyme in bacterial purine metabolism, plays an essential role in the biosynthesis of guanine nucleotides and shows promise as a target for ...Inosine 5'-monophosphate dehydrogenase (IMPDH), a key enzyme in bacterial purine metabolism, plays an essential role in the biosynthesis of guanine nucleotides and shows promise as a target for antimicrobial drug development. Despite its significance, the conformational dynamics and substrate-induced structural changes in bacterial IMPDH remain poorly understood, particularly with respect to its octameric assembly. Using cryo-EM, we present full-length structures of IMPDH from Mycobacterium smegmatis (MsmIMPDH) captured in a reaction intermediate state, revealing conformational changes upon substrate binding. The structures feature resolved flexible loops that coordinate the binding of the substrate, the cofactor, and the K ion. Our structural analysis identifies a novel octamerization interface unique to MsmIMPDH. Additionally, a previously unobserved barrel-like density suggests potential self-interactions within the C-terminal regions, hinting at a regulatory mechanism tied to assembly and function of the enzyme. These data provide insights into substrate-induced conformational dynamics and novel interaction interfaces in MsmIMPDH, potentially informing the development of IMPDH-targeted drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_52559.map.gz emd_52559.map.gz | 136 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-52559-v30.xml emd-52559-v30.xml emd-52559.xml emd-52559.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_52559_fsc.xml emd_52559_fsc.xml | 14.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_52559.png emd_52559.png | 131.4 KB | ||
| Masks |  emd_52559_msk_1.map emd_52559_msk_1.map | 282.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-52559.cif.gz emd-52559.cif.gz | 7.1 KB | ||
| Others |  emd_52559_additional_1.map.gz emd_52559_additional_1.map.gz emd_52559_half_map_1.map.gz emd_52559_half_map_1.map.gz emd_52559_half_map_2.map.gz emd_52559_half_map_2.map.gz | 263.1 MB 225.3 MB 225.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-52559 http://ftp.pdbj.org/pub/emdb/structures/EMD-52559 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-52559 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-52559 | HTTPS FTP |
-Related structure data
| Related structure data |  9i0kMC  9i0lC  9i0mC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_52559.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_52559.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LocScale primary viewing map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8336 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_52559_msk_1.map emd_52559_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Original map used for refinement and validation
| File | emd_52559_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Original map used for refinement and validation | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_52559_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_52559_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Octameric assembly of inosine monophosphate dehydrogenase reactio...
| Entire | Name: Octameric assembly of inosine monophosphate dehydrogenase reaction intermediate; E-XMP* in complex with NAD+ |
|---|---|
| Components |
|
-Supramolecule #1: Octameric assembly of inosine monophosphate dehydrogenase reactio...
| Supramolecule | Name: Octameric assembly of inosine monophosphate dehydrogenase reaction intermediate; E-XMP* in complex with NAD+ type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
-Macromolecule #1: Inosine-5'-monophosphate dehydrogenase
| Macromolecule | Name: Inosine-5'-monophosphate dehydrogenase / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO / EC number: IMP dehydrogenase |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) / Strain: MC2 155 Mycolicibacterium smegmatis MC2 155 (bacteria) / Strain: MC2 155 |
| Molecular weight | Theoretical: 53.388988 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSIAESSVPI AVPVPTGGDD PTKVAMLGLT FDDVLLLPAA SDVVPATADT SSQLTKRIRL RVPLVSSAMD TVTESRMAIA MARAGGMGV LHRNLPVAEQ AGQVETVKRS EAGMVTDPVT CSPDNTLAEV DAMCARFRIS GLPVVDDTGE LVGIITNRDM R FEVDQSKP ...String: MSIAESSVPI AVPVPTGGDD PTKVAMLGLT FDDVLLLPAA SDVVPATADT SSQLTKRIRL RVPLVSSAMD TVTESRMAIA MARAGGMGV LHRNLPVAEQ AGQVETVKRS EAGMVTDPVT CSPDNTLAEV DAMCARFRIS GLPVVDDTGE LVGIITNRDM R FEVDQSKP VSEVMTKAPL ITAKEGVSAE AALGLLRRHK IEKLPIVDGH GKLTGLITVK DFVKTEQFPL STKDSDGRLL VG AAVGVGD DAWTRAMTLV DAGVDVLIVD TAHAHNRGVL DMVSRLKQAV GERVDVVGGN VATRAAAAAL VEAGADAVKV GVG PGSICT TRVVAGVGAP QITAILEAVA ACKPYGVPVI ADGGLQYSGD IAKALAAGAS TAMLGSLLAG TAESPGELIF VNGK QFKSY RGMGSLGAMQ GRGAAKSYSK DRYFQDDVLS EDKLVPEGIE GRVPFRGPLG TVIHQLTGGL RAAMGYTGSA TIEQL QQAQ FVQITAAGLK ESHPHDITMT VEAPNYYTR UniProtKB: Inosine-5'-monophosphate dehydrogenase |
-Macromolecule #2: INOSINIC ACID
| Macromolecule | Name: INOSINIC ACID / type: ligand / ID: 2 / Number of copies: 8 / Formula: IMP |
|---|---|
| Molecular weight | Theoretical: 348.206 Da |
| Chemical component information | 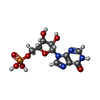 ChemComp-I: |
-Macromolecule #3: NICOTINAMIDE-ADENINE-DINUCLEOTIDE
| Macromolecule | Name: NICOTINAMIDE-ADENINE-DINUCLEOTIDE / type: ligand / ID: 3 / Number of copies: 8 / Formula: NAD |
|---|---|
| Molecular weight | Theoretical: 663.425 Da |
| Chemical component information |  ChemComp-NAD: |
-Macromolecule #4: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 4 / Number of copies: 8 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 80 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
Details: 50 mM HEPES (pH 7.5), 200 mM KCl, 5 mM DTT, 32 mM MgCl2 Ligand: 10 mM ATP + 10 mM IMP + 10 mM NAD | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 8691 / Average exposure time: 2.0 sec. / Average electron dose: 40.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 165000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Initial fitting was done in UCSF ChimeraX. Model refinement was done by iterative cycles of manual fitting with Coot and ISOLDE and automated fitting with phenix.real_space_refine. |
| Refinement | Protocol: RIGID BODY FIT / Overall B value: 37.48 / Target criteria: CC coefficient |
| Output model |  PDB-9i0k: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)























































