+Search query
-Structure paper
| Title | Conformational landscape of the mycobacterial inosine 5'-monophosphate dehydrogenase octamerization interface. |
|---|---|
| Journal, issue, pages | J Struct Biol, Vol. 217, Issue 2, Page 108198, Year 2025 |
| Publish date | Mar 17, 2025 |
 Authors Authors | Ondřej Bulvas / Zdeněk Knejzlík / Anatolij Filimoněnko / Tomáš Kouba / Iva Pichová /  |
| PubMed Abstract | Inosine 5'-monophosphate dehydrogenase (IMPDH), a key enzyme in bacterial purine metabolism, plays an essential role in the biosynthesis of guanine nucleotides and shows promise as a target for ...Inosine 5'-monophosphate dehydrogenase (IMPDH), a key enzyme in bacterial purine metabolism, plays an essential role in the biosynthesis of guanine nucleotides and shows promise as a target for antimicrobial drug development. Despite its significance, the conformational dynamics and substrate-induced structural changes in bacterial IMPDH remain poorly understood, particularly with respect to its octameric assembly. Using cryo-EM, we present full-length structures of IMPDH from Mycobacterium smegmatis (MsmIMPDH) captured in a reaction intermediate state, revealing conformational changes upon substrate binding. The structures feature resolved flexible loops that coordinate the binding of the substrate, the cofactor, and the K ion. Our structural analysis identifies a novel octamerization interface unique to MsmIMPDH. Additionally, a previously unobserved barrel-like density suggests potential self-interactions within the C-terminal regions, hinting at a regulatory mechanism tied to assembly and function of the enzyme. These data provide insights into substrate-induced conformational dynamics and novel interaction interfaces in MsmIMPDH, potentially informing the development of IMPDH-targeted drugs. |
 External links External links |  J Struct Biol / J Struct Biol /  PubMed:40107326 PubMed:40107326 |
| Methods | EM (single particle) |
| Resolution | 2.39 - 3.01 Å |
| Structure data | EMDB-52559, PDB-9i0k: EMDB-52560, PDB-9i0l: EMDB-52561, PDB-9i0m: |
| Chemicals | 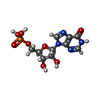 ChemComp-IMP:  ChemComp-NAD:  ChemComp-K:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords | OXIDOREDUCTASE / Octamer / reaction intermediate / Purine metabolism / IMPDH / ligand complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers









 mycolicibacterium smegmatis mc2 155 (bacteria)
mycolicibacterium smegmatis mc2 155 (bacteria)