[English] 日本語
 Yorodumi
Yorodumi- EMDB-4943: In situ structure of a pentameric IgM complex bound to complement... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4943 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ structure of a pentameric IgM complex bound to complement components C1 and one molecule of C4b | |||||||||
 Map data Map data | Pentameric IgM with C1 and one copy of C4b | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
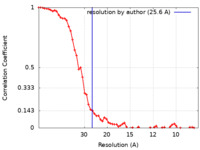
| Method | subtomogram averaging / cryo EM / Resolution: 25.6 Å | |||||||||
 Authors Authors | Sharp TH | |||||||||
| Funding support |  Netherlands, 1 items Netherlands, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2019 Journal: Proc Natl Acad Sci U S A / Year: 2019Title: Insights into IgM-mediated complement activation based on in situ structures of IgM-C1-C4b. Authors: Thomas H Sharp / Aimee L Boyle / Christoph A Diebolder / Alexander Kros / Abraham J Koster / Piet Gros /  Abstract: Antigen binding by serum Ig-M (IgM) protects against microbial infections and helps to prevent autoimmunity, but causes life-threatening diseases when mistargeted. How antigen-bound IgM activates ...Antigen binding by serum Ig-M (IgM) protects against microbial infections and helps to prevent autoimmunity, but causes life-threatening diseases when mistargeted. How antigen-bound IgM activates complement-immune responses remains unclear. We present cryoelectron tomography structures of IgM, C1, and C4b complexes formed on antigen-bearing lipid membranes by normal human serum at 4 °C. The IgM-C1-C4b complexes revealed C4b product release as the temperature-limiting step in complement activation. Both IgM hexamers and pentamers adopted hexagonal, dome-shaped structures with Fab pairs, dimerized by hinge domains, bound to surface antigens that support a platform of Fc regions. C1 binds IgM through widely spread C1q-collagen helices, with C1r proteases pointing outward and C1s bending downward and interacting with surface-attached C4b, which further interacts with the adjacent IgM-Fab and globular C1q-recognition unit. Based on these data, we present mechanistic models for antibody-mediated, C1q-transmitted activation of C1 and for C4b deposition, while further conformational rearrangements are required to form C3 convertases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4943.map.gz emd_4943.map.gz | 14.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4943-v30.xml emd-4943-v30.xml emd-4943.xml emd-4943.xml | 12.4 KB 12.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4943_fsc.xml emd_4943_fsc.xml | 6.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_4943.png emd_4943.png | 153.3 KB | ||
| Masks |  emd_4943_msk_1.map emd_4943_msk_1.map | 15.6 MB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4943 http://ftp.pdbj.org/pub/emdb/structures/EMD-4943 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4943 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4943 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4943.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4943.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Pentameric IgM with C1 and one copy of C4b | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4943_msk_1.map emd_4943_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : pentameric IgM-C1-C4b1
| Entire | Name: pentameric IgM-C1-C4b1 |
|---|---|
| Components |
|
-Supramolecule #1: pentameric IgM-C1-C4b1
| Supramolecule | Name: pentameric IgM-C1-C4b1 / type: complex / ID: 1 / Parent: 0 Details: Map contains a complex of pentameric IgM, complement component C1 and one molecule of complement component C4b |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: Human serum Homo sapiens (human) / Organ: Human serum |
| Molecular weight | Theoretical: 1.931 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Component - Formula: PBS / Component - Name: PBS |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 277.15 K / Instrument: LEICA EM GP |
| Details | Antigenic liposomes + IgM + Normal human serum |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 2.4 sec. / Average electron dose: 1.48 e/Å2 Details: Exposures of 2.4 sec were dose-fractionated into 6 movie frames per tilt angle. The total dose for each tilt series was 80 e-/A^2. Focusing to -300 nm was performed before each image ...Details: Exposures of 2.4 sec were dose-fractionated into 6 movie frames per tilt angle. The total dose for each tilt series was 80 e-/A^2. Focusing to -300 nm was performed before each image acquisition using a low-dose routine |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 33000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)