[English] 日本語
 Yorodumi
Yorodumi- EMDB-4722: Structure of a soluble domain of adenylyl cyclase bound to an act... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4722 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a soluble domain of adenylyl cyclase bound to an activatedstimulatory G protein | |||||||||
 Map data Map data | Soluble domain of AC9 bound to GalphaS | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane protein / adenylyl cyclase / G protein / occluded state | |||||||||
| Function / homology |  Function and homology information Function and homology informationAdenylate cyclase activating pathway / Adenylate cyclase inhibitory pathway / PKA activation / adenylate cyclase / sensory perception of chemical stimulus / Hedgehog 'off' state / mu-type opioid receptor binding / cAMP biosynthetic process / corticotropin-releasing hormone receptor 1 binding / adenylate cyclase activity ...Adenylate cyclase activating pathway / Adenylate cyclase inhibitory pathway / PKA activation / adenylate cyclase / sensory perception of chemical stimulus / Hedgehog 'off' state / mu-type opioid receptor binding / cAMP biosynthetic process / corticotropin-releasing hormone receptor 1 binding / adenylate cyclase activity / beta-2 adrenergic receptor binding / G alpha (z) signalling events / D1 dopamine receptor binding / adenylate cyclase-activating adrenergic receptor signaling pathway / insulin-like growth factor receptor binding / ionotropic glutamate receptor binding / adenylate cyclase activator activity / G-protein beta/gamma-subunit complex binding / adenylate cyclase-activating G protein-coupled receptor signaling pathway / adenylate cyclase-activating dopamine receptor signaling pathway / heterotrimeric G-protein complex / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / in utero embryonic development / intracellular signal transduction / ciliary basal body / GTPase activity / GTP binding / ATP binding / metal ion binding / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
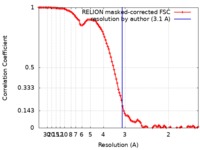
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Korkhov VM / Qi C | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2019 Journal: Science / Year: 2019Title: The structure of a membrane adenylyl cyclase bound to an activated stimulatory G protein. Authors: Chao Qi / Simona Sorrentino / Ohad Medalia / Volodymyr M Korkhov /   Abstract: Membrane-integral adenylyl cyclases (ACs) are key enzymes in mammalian heterotrimeric GTP-binding protein (G protein)-dependent signal transduction, which is important in many cellular processes. ...Membrane-integral adenylyl cyclases (ACs) are key enzymes in mammalian heterotrimeric GTP-binding protein (G protein)-dependent signal transduction, which is important in many cellular processes. Signals received by the G protein-coupled receptors are conveyed to ACs through G proteins to modulate the levels of cellular cyclic adenosine monophosphate (cAMP). Here, we describe the cryo-electron microscopy structure of the bovine membrane AC9 bound to an activated G protein αs subunit at 3.4-angstrom resolution. The structure reveals the organization of the membrane domain and helical domain that spans between the membrane and catalytic domains of AC9. The carboxyl-terminal extension of the catalytic domain occludes both the catalytic and the allosteric sites of AC9, inducing a conformation distinct from the substrate- and activator-bound state, suggesting a regulatory role in cAMP production. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4722.map.gz emd_4722.map.gz | 8.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4722-v30.xml emd-4722-v30.xml emd-4722.xml emd-4722.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4722_fsc.xml emd_4722_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_4722.png emd_4722.png | 66.3 KB | ||
| Filedesc metadata |  emd-4722.cif.gz emd-4722.cif.gz | 7.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4722 http://ftp.pdbj.org/pub/emdb/structures/EMD-4722 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4722 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4722 | HTTPS FTP |
-Related structure data
| Related structure data |  6r4pMC  4719C  4721C  4723C  4724C  4725C  4726C  6r3qC  6r4oC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4722.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4722.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Soluble domain of AC9 bound to GalphaS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.814 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
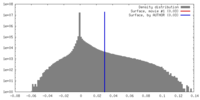
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of adenylyl cyclase AC9 with G protein subunit Galphas
| Entire | Name: Complex of adenylyl cyclase AC9 with G protein subunit Galphas |
|---|---|
| Components |
|
-Supramolecule #1: Complex of adenylyl cyclase AC9 with G protein subunit Galphas
| Supramolecule | Name: Complex of adenylyl cyclase AC9 with G protein subunit Galphas type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|
-Supramolecule #2: Adenylate cyclase 9
| Supramolecule | Name: Adenylate cyclase 9 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Supramolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Adenylate cyclase 9
| Macromolecule | Name: Adenylate cyclase 9 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 182.404422 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASPPHQQLL QHHSTEVSCD SSGDSNSVRV RINPKQPSSN SHPKHCKYSI SSSCSSSGDS GGVPRRMGAG GRLRRRKKLP QLFERASSR WWDPKFDSVN LEEACMERCF PQTQRRFRYA LFYIGFACLL WSIYFGVHMK SKLIVMVAPA LCFLVVCVGF F LFTFTKLY ...String: MASPPHQQLL QHHSTEVSCD SSGDSNSVRV RINPKQPSSN SHPKHCKYSI SSSCSSSGDS GGVPRRMGAG GRLRRRKKLP QLFERASSR WWDPKFDSVN LEEACMERCF PQTQRRFRYA LFYIGFACLL WSIYFGVHMK SKLIVMVAPA LCFLVVCVGF F LFTFTKLY ARHYVWTSLV LTLLVFALTL AAQFQVLTPL SGRVDNFNHT RAARPTDTCL SQVGSFSMCI EVLFLLYTVM HL PLYLSLI LGVAYSVLFE TFGYHFQDEA CFASPGAEAL HWELLSRALL HLCIHAIGIH LFIMSQVRSR STFLKVGQSI MHG KDLEVE KALKERMIHS VMPRIIADDL MKQGDEESEN SVKRHATSSP KNRKKKSSIQ KAPIAFRPFK MQQIEEVSIL FADI VGFTK MSANKSAHAL VGLLNDLFGR FDRLCEETKC EKISTLGDCY YCVAGCPEPR ADHAYCCIEM GLGMIRAIEQ FCQEK KEMV NMRVGVHTGT VLCGILGMRR FKFDVWSNDV NLANLMEQLG VAGKVHISEA TAKYLDDRYE MEDGKVTERL GQSVVA DQL KGLKTYLIAG QRAKESHCSC SEALLSGFEV LDGSRVSSGP RGQGTASPGS VSDLAQTVKT FDNLKTCPSC GITFTPK PE AGAEGGAVQN GCQEEPKNSA KASGGPSSKT QNGLLSPPPE EKLTNSQTSL CEILQEKGRW AGVSLDQSAL LPLRFKNI R EKTDAHFVDV IKEDSLMKDY FFKPPINQFS LNFLDPELER AYRTSYQEEV VKSSPVRTFA SATFSSLLDV LLSTTVFLI LSITCFLRYG AASTPPPPAA LAVFGAALLL EILSLVVSVR MVFFLEDVMT CTKRLLEWIA GWLPRHFIGA ILVSLPALAV YSHVTSEFE TNIHSTMFTG SAVLTAVVQY CNFCQLSSWM RSSLATVVGA GPLLLLLYVS LCPDSSTVIS HLDAVQNFSS T RKLCNASL PHDGRSPASL IGQEVILVFF LLLLLVWFLN REFEVSYRLH YHGDVEADLH RTKIQSMRDQ ADWLLRNIIP YH VAEQLKV SQTYSKNHDS GGVIFASIVN FSEFYEENYE GGKECYRVLN ELIGDFDELL SKPDYSSIEK IKTIGATYMA ASG LNATQC RDGSHPQEHL QILFEFAKEM MRVVDDFNNN MLWFNFKLRV GFNHGPLTAG VIGTTKLLYD IWGDTVNIAS RMDT TGVEC RIQVSEESYR VLSKMGYEFD YRGTVNVKGK GQMKTYLYPK CTDSGLVPQH QLSISPDIRV QVDGSIGRSP TDEIA SLVP SVQNPDQVPP GSENNAQTRD AHPSAKRPWK EPVRAEERCR FGKAIEKSDC EEVGMEEANE LTKLNVSERA AAALEV LFQ GPGGVSKGEE LFTGVVPILV ELDGDVNGHK FSVSGEGEGD ATYGKLTLKF ICTTGKLPVP WPTLVTTFGY GLQCFAR YP DHMKQHDFFK SAMPEGYVQE RTIFFKDDGN YKTRAEVKFE GDTLVNRIEL KGIDFKEDGN ILGHKLEYNY NSHNVYIM A DKQKNGIKVN FKIRHNIEDG SVQLADHYQQ NTPIGDGPVL LPDNHYLSYQ SALSKDPNEK RDHMVLLEFV TAAGITLGM DELYKAASAW SHPQFEKGGG SGGGSGGSAW SHPQFEK UniProtKB: Adenylate cyclase type 9 |
-Macromolecule #2: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 46.932727 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKSTIVKQM RILHVNGFNG GEGGEEDPNA KSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE ...String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKSTIVKQM RILHVNGFNG GEGGEEDPNA KSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE YQLIDCAQYF LDKIDVIKQD DYVPSDQDLL RCRVLTSGIF ETKFQVDKVN FHMFDVGGQR DERRKWIQCF ND VTAIIFV VASSSYNMVI REDNQTNRLQ EALNLFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK IEDYFPEFAR YTT PEDATP EPGEDPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCA VDTENIRRVF NDCRDIIQRM HLRQYELLGG HHHH HHHH UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #3: 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE
| Macromolecule | Name: 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE / type: ligand / ID: 3 / Number of copies: 1 / Formula: GSP |
|---|---|
| Molecular weight | Theoretical: 539.246 Da |
| Chemical component information |  ChemComp-GSP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 5817 / Average exposure time: 8.0 sec. / Average electron dose: 47.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.75 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 86.68 / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-6r4p: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)