[English] 日本語
 Yorodumi
Yorodumi- EMDB-37975: Human FL Metabotropic glutamate receptor 5, mGlu5-5M with quisqua... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human FL Metabotropic glutamate receptor 5, mGlu5-5M with quisqualate and VU0424465, conformer 2 | ||||||||||||
 Map data Map data | combined sharpened map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | G-PROTEIN COUPLED RECEPTORS / SIGNAL TRANSDUCTION / METABOTROPIC GLUTAMATE RECEPTOR / Agonist / active state / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsensory perception of hot stimulus / A2A adenosine receptor binding / : / negative regulation of dendritic spine morphogenesis / G protein-coupled receptor activity involved in regulation of postsynaptic membrane potential / adenylate cyclase inhibiting G protein-coupled glutamate receptor activity / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / operant conditioning / protein localization to nuclear inner membrane / positive regulation of cellular response to hypoxia ...sensory perception of hot stimulus / A2A adenosine receptor binding / : / negative regulation of dendritic spine morphogenesis / G protein-coupled receptor activity involved in regulation of postsynaptic membrane potential / adenylate cyclase inhibiting G protein-coupled glutamate receptor activity / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / operant conditioning / protein localization to nuclear inner membrane / positive regulation of cellular response to hypoxia / positive regulation of long-term neuronal synaptic plasticity / desensitization of G protein-coupled receptor signaling pathway / positive regulation of sensory perception of pain / negative regulation of excitatory postsynaptic potential / positive regulation of dopamine secretion / G protein-coupled glutamate receptor signaling pathway / Class C/3 (Metabotropic glutamate/pheromone receptors) / positive regulation of neural precursor cell proliferation / glutamate receptor activity / nuclear inner membrane / Neurexins and neuroligins / astrocyte projection / response to corticosterone / response to morphine / temperature homeostasis / protein tyrosine kinase activator activity / conditioned place preference / regulation of synaptic transmission, glutamatergic / regulation of long-term synaptic depression / positive regulation of calcium-mediated signaling / protein tyrosine kinase binding / dendritic shaft / response to amphetamine / locomotory behavior / synapse organization / postsynaptic density membrane / G protein-coupled receptor activity / cognition / Schaffer collateral - CA1 synapse / cellular response to amyloid-beta / positive regulation of cytosolic calcium ion concentration / G alpha (q) signalling events / dendritic spine / response to ethanol / chemical synaptic transmission / learning or memory / positive regulation of MAPK cascade / response to antibiotic / neuronal cell body / dendrite / regulation of DNA-templated transcription / glutamatergic synapse / identical protein binding / plasma membrane / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||
 Authors Authors | Lebon G / Cannone G / Vinothkumar KR | ||||||||||||
| Funding support |  India, India,  France, 3 items France, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Conformational diversity in class C GPCR positive allosteric modulation. Authors: Giuseppe Cannone / Ludovic Berto / Fanny Malhaire / Gavin Ferguson / Aurelien Fouillen / Stéphanie Balor / Joan Font-Ingles / Amadeu Llebaria / Cyril Goudet / Abhay Kotecha / Vinothkumar K ...Authors: Giuseppe Cannone / Ludovic Berto / Fanny Malhaire / Gavin Ferguson / Aurelien Fouillen / Stéphanie Balor / Joan Font-Ingles / Amadeu Llebaria / Cyril Goudet / Abhay Kotecha / Vinothkumar K R / Guillaume Lebon /      Abstract: The metabotropic glutamate receptors (mGlus) are class C G protein-coupled receptors (GPCR) that form obligate dimers activated by the major excitatory neurotransmitter L-glutamate. The architecture ...The metabotropic glutamate receptors (mGlus) are class C G protein-coupled receptors (GPCR) that form obligate dimers activated by the major excitatory neurotransmitter L-glutamate. The architecture of mGlu receptor comprises an extracellular Venus-Fly Trap domain (VFT) connected to the transmembrane domain (7TM) through a Cysteine-Rich Domain (CRD). The binding of L-glutamate in the VFTs and subsequent conformational change results in the signal being transmitted to the 7TM inducing G protein binding and activation. The mGlu receptors signal transduction can be allosterically potentiated by positive allosteric modulators (PAMs) binding to the 7TMs, which are of therapeutic interest in various neurological disorders. Here, we report the cryoEM structures of metabotropic glutamate receptor 5 (mGlu) purified with three chemically and pharmacologically distinct PAMs. We find that the PAMs modulate the receptor equilibrium through their different binding modes, revealing how their interactions in the 7TMs impact the mGlu receptor conformational landscape and function. In addition, we identified a PAM-free but agonist-bound intermediate state that also reveals interactions mediated by intracellular loop 2. The activation of mGlu receptor is a multi-step process in which the binding of the PAMs in the 7TM modulates the equilibrium towards the active state. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37975.map.gz emd_37975.map.gz | 59.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37975-v30.xml emd-37975-v30.xml emd-37975.xml emd-37975.xml | 25 KB 25 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37975_fsc.xml emd_37975_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_37975.png emd_37975.png | 86 KB | ||
| Filedesc metadata |  emd-37975.cif.gz emd-37975.cif.gz | 8.1 KB | ||
| Others |  emd_37975_additional_1.map.gz emd_37975_additional_1.map.gz emd_37975_half_map_1.map.gz emd_37975_half_map_1.map.gz emd_37975_half_map_2.map.gz emd_37975_half_map_2.map.gz | 56.6 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37975 http://ftp.pdbj.org/pub/emdb/structures/EMD-37975 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37975 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37975 | HTTPS FTP |
-Related structure data
| Related structure data |  8x0dMC  8x0bC  8x0cC  8x0eC  8x0fC  8x0gC  8x0hC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37975.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37975.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | combined sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02259 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: DeepEMhancer map used during model building
| File | emd_37975_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer map used during model building | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: One of the half maps
| File | emd_37975_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | One of the half maps | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: One of the half maps
| File | emd_37975_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | One of the half maps | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human FL Metabotropic glutamate receptor 5, mGlu5-5M
| Entire | Name: Human FL Metabotropic glutamate receptor 5, mGlu5-5M |
|---|---|
| Components |
|
-Supramolecule #1: Human FL Metabotropic glutamate receptor 5, mGlu5-5M
| Supramolecule | Name: Human FL Metabotropic glutamate receptor 5, mGlu5-5M / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Receptor purified in detergent micelles with agonist quisqualate and PAM |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 200 KDa |
-Macromolecule #1: Metabotropic glutamate receptor 5
| Macromolecule | Name: Metabotropic glutamate receptor 5 / type: protein_or_peptide / ID: 1 Details: The construct has tags and protease cleavage site at its N-term (DYKDDDDKHHHHHHHHHHLEVLFQGP) and when compared to uniprot it starts at 21 and ends at 856. For CryoEM, the tag is cleaved and ...Details: The construct has tags and protease cleavage site at its N-term (DYKDDDDKHHHHHHHHHHLEVLFQGP) and when compared to uniprot it starts at 21 and ends at 856. For CryoEM, the tag is cleaved and the protein used for experiment starts at QSSE but residues starting RR have been modeled. There are 5 thermostabilising mutations (T742A, S753A, T777A, I799A, A813L). Residue N445 is mutated to remove glycosylation. An additional mutation H350L is engineered for a nanobody to bind. Each monomer has one sugar modeled and 10 disulfide bonds. Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 93.691492 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSSERRVVAH MPGDIIIGAL FSVHHQPTVD KVHERKCGAV REQYGIQRVE AMLHTLERIN SDPTLLPNIT LGCEIRDSCW HSAVALEQS IEFIRDSLIS SEEEEGLVRC VDGSSSSFRS KKPIVGVIGP GSSSVAIQVQ NLLQLFNIPQ IAYSATSMDL S DKTLFKYF ...String: QSSERRVVAH MPGDIIIGAL FSVHHQPTVD KVHERKCGAV REQYGIQRVE AMLHTLERIN SDPTLLPNIT LGCEIRDSCW HSAVALEQS IEFIRDSLIS SEEEEGLVRC VDGSSSSFRS KKPIVGVIGP GSSSVAIQVQ NLLQLFNIPQ IAYSATSMDL S DKTLFKYF MRVVPSDAQQ ARAMVDIVKR YNWTYVSAVH TEGNYGESGM EAFKDMSAKE GICIAHSYKI YSNAGEQSFD KL LKKLTSH LPKARVVACF CEGMTVRGLL MAMRRLGLAG EFLLLGSDGW ADRYDVTDGY QREAVGGITI KLQSPDVKWF DDY YLKLRP ETNLRNPWFQ EFWQHRFQCR LEGFPQENSK YNKTCNSSLT LKTHHVQDSK MGFVINAIYS MAYGLHNMQM SLCP GYAGL CDAMKPIDGR KLLESLMKTA FTGVSGDTIL FDENGDSPGR YEIMNFKEMG KDYFDYINVG SWDNGELKMD DDEVW SKKS NIIRSVCSEP CEKGQIKVIR KGEVSCCWTC TPCKENEYVF DEYTCKACQL GSWPTDDLTG CDLIPVQYLR WGDPEP IAA VVFACLGLLA TLFVTVVFII YRDTPVVKSS SRELCYIILA GICLGYLCTF CLIAKPKQIY CYLQRIGIGL SPAMSYS AL VTKTNRIARI LAGSKKKICT KKPRFMSACA QLVIAFILIC IQLGIIVALF IMEPPDIMHD YPSIREVYLI CNTTNLGV V APLGYNGLLI LACTFYAFKT RNVPANFNEA KYIAFAMYTT CIIWLAFVPI YFGSNYKAIT MCFSVSLSAT VLLGCMFVP KVYIILAKPE RNVRSAFTTS TVVRMHVGDG KSSSAA UniProtKB: Metabotropic glutamate receptor 5 |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #3: (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID
| Macromolecule | Name: (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID type: ligand / ID: 3 / Number of copies: 2 / Formula: QUS |
|---|---|
| Molecular weight | Theoretical: 189.126 Da |
| Chemical component information | 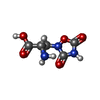 ChemComp-QUS: |
-Macromolecule #4: VU0424465
| Macromolecule | Name: VU0424465 / type: ligand / ID: 4 / Number of copies: 2 / Formula: XQT |
|---|---|
| Molecular weight | Theoretical: 326.365 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV / Details: Blotting time of 5 seconds and blot force of 10. | ||||||||||
| Details | Receptor is purified in detergent micelles and monodisperse. The receptor is purified with 10 uM Quisqualate and 10 uM PAM VU0424465 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | Images were collected with Aberration-free image shift and Fringe-free imaging with a Selectris energy filter in EFTEM mode |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 19115 / Average exposure time: 4.9 sec. / Average electron dose: 50.0 e/Å2 / Details: Electron flux was 5.4 /e/A2/s. No. of frames 1566 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 192572 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Source name: Other / Chain - Initial model type: other Details: The model which is being curated was used as initial template |
|---|---|
| Refinement | Space: RECIPROCAL / Protocol: OTHER / Overall B value: 95.3 |
| Output model |  PDB-8x0d: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)














































