+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ATG2A-WIPI4 complex | |||||||||
 Map data Map data | Half map A of ATG2A-WIPI4 complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ATG2A / single particle cryo-EM / peripheral membrane proteins / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationphagophore / lipid transfer activity / organelle membrane contact site / glycophagy / positive regulation of autophagosome assembly / phagophore assembly site membrane / autophagy of mitochondrion / pexophagy / piecemeal microautophagy of the nucleus / phosphatidylinositol-3-phosphate binding ...phagophore / lipid transfer activity / organelle membrane contact site / glycophagy / positive regulation of autophagosome assembly / phagophore assembly site membrane / autophagy of mitochondrion / pexophagy / piecemeal microautophagy of the nucleus / phosphatidylinositol-3-phosphate binding / phagophore assembly site / reticulophagy / autophagosome assembly / protein-membrane adaptor activity / positive regulation of autophagy / lipid droplet / endoplasmic reticulum membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
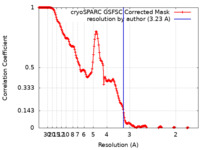
| Method | single particle reconstruction / cryo EM / Resolution: 3.23 Å | |||||||||
 Authors Authors | Wang Y / Stjepanovic G | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2025 Journal: Nat Struct Mol Biol / Year: 2025Title: Structural basis for lipid transfer by the ATG2A-ATG9A complex. Authors: Yang Wang / Selma Dahmane / Rujuan Ti / Xinyi Mai / Lizhe Zhu / Lars-Anders Carlson / Goran Stjepanovic /   Abstract: Autophagy is characterized by the formation of double-membrane vesicles called autophagosomes. Autophagy-related proteins (ATGs) 2A and 9A have an essential role in autophagy by mediating lipid ...Autophagy is characterized by the formation of double-membrane vesicles called autophagosomes. Autophagy-related proteins (ATGs) 2A and 9A have an essential role in autophagy by mediating lipid transfer and re-equilibration between membranes for autophagosome formation. Here we report the cryo-electron microscopy structures of human ATG2A in complex with WD-repeat protein interacting with phosphoinositides 4 (WIPI4) at 3.2 Å and the ATG2A-WIPI4-ATG9A complex at 7 Å global resolution. On the basis of molecular dynamics simulations, we propose a mechanism of lipid extraction from the donor membranes. Our analysis revealed 3:1 stoichiometry of the ATG9A-ATG2A complex, directly aligning the ATG9A lateral pore with ATG2A lipid transfer cavity, and an interaction of the ATG9A trimer with both the N-terminal and the C-terminal tip of rod-shaped ATG2A. Cryo-electron tomography of ATG2A liposome-binding states showed that ATG2A tethers lipid vesicles at different orientations. In summary, this study provides a molecular basis for the growth of the phagophore membrane and lends structural insights into spatially coupled lipid transport and re-equilibration during autophagosome formation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37087.map.gz emd_37087.map.gz | 483.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37087-v30.xml emd-37087-v30.xml emd-37087.xml emd-37087.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37087_fsc.xml emd_37087_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_37087.png emd_37087.png | 55.5 KB | ||
| Filedesc metadata |  emd-37087.cif.gz emd-37087.cif.gz | 6.5 KB | ||
| Others |  emd_37087_half_map_1.map.gz emd_37087_half_map_1.map.gz emd_37087_half_map_2.map.gz emd_37087_half_map_2.map.gz | 475.8 MB 475.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37087 http://ftp.pdbj.org/pub/emdb/structures/EMD-37087 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37087 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37087 | HTTPS FTP |
-Related structure data
| Related structure data |  8kbyMC  8kbxC  8kbzC  8kc3C  8y1lC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_37087.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37087.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of ATG2A-WIPI4 complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
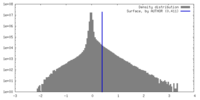
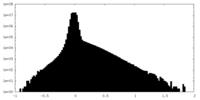
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map B of ATG2A-WIPI4 complex
| File | emd_37087_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of ATG2A-WIPI4 complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
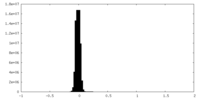
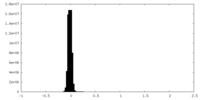
| Density Histograms |
-Half map: Half map B of ATG2A-WIPI4 complex
| File | emd_37087_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of ATG2A-WIPI4 complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ATG2A
| Entire | Name: ATG2A |
|---|---|
| Components |
|
-Supramolecule #1: ATG2A
| Supramolecule | Name: ATG2A / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 213 KDa |
-Macromolecule #1: Autophagy-related protein 2 homolog A
| Macromolecule | Name: Autophagy-related protein 2 homolog A / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 213.100281 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSRWLWPWSN CVKERVCRYL LHHYLGHFFQ EHLSLDQLSL DLYKGSVALR DIHLEIWSVN EVLESMESPL ELVEGFVGSI EVAVPWAAL LTDHCTVRVS GLQLTLQPRR GPAPGAADSQ SWASCMTTSL QLAQECLRDG LPEPSEPPQP LEGLEMFAQT I ETVLRRIK ...String: MSRWLWPWSN CVKERVCRYL LHHYLGHFFQ EHLSLDQLSL DLYKGSVALR DIHLEIWSVN EVLESMESPL ELVEGFVGSI EVAVPWAAL LTDHCTVRVS GLQLTLQPRR GPAPGAADSQ SWASCMTTSL QLAQECLRDG LPEPSEPPQP LEGLEMFAQT I ETVLRRIK VTFLDTVVRV EHSPGDGERG VAVEVRVQRL EYCDEAVRDP SQAPPVDVHQ PPAFLHKLLQ LAGVRLHYEE LP AQEEPPE PPLQIGSCSG YMELMVKLKQ NEAFPGPKLE VAGQLGSLHL LLTPRQLQQL QELLSAVSLT DHEGLADKLN KSR PLGAED LWLIEQDLNQ QLQAGAVAEP LSPDPLTNPL LNLDNTDLFF SMAGLTSSVA SALSELSLSD VDLASSVRSD MASR RLSAQ AHPAGKMAPN PLLDTMRPDS LLKMTLGGVT LTLLQTSAPS SGPPDLATHF FTEFDATKDG PFGSRDFHHL RPRFQ RACP CSHVRLTGTA VQLSWELRTG SRGRRTTSME VHFGQLEVLE CLWPRGTSEP EYTEILTFPG TLGSQASARP CAHLRH TQI LRRVPKSRPR RSVACHCHSE LALDLANFQA DVELGALDRL AALLRLATVP AEPPAGLLTE PLPAMEQQTV FRLSAPR AT LRLRFPIADL RPEPDPWAGQ AVRAEQLRLE LSEPQFRSEL SSGPGPPVPT HLELTCSDLH GIYEDGGKPP VPCLRVSK A LDPKSTGRKY FLPQVVVTVN PQSSSTQWEV APEKGEELEL SVESPCELRE PEPSPFSSKR TMYETEEMVI PGDPEEMRT FQSRTLALSR CSLEVILPSV HIFLPSKEVY ESIYNRINND LLMWEPADLL PTPDPAAQPS GFPGPSGFWH DSFKMCKSAF KLANCFDLT PDSDSDDEDA HFFSVGASGG PQAAAPEAPS LHLQSTFSTL VTVLKGRITA LCETKDEGGK RLEAVHGELV L DMEHGTLF SVSQYCGQPG LGYFCLEAEK ATLYHRAAVD DYPLPSHLDL PSFAPPAQLA PTIYPSEEGV TERGASGRKG QG RGPHMLS TAVRIHLDPH KNVKEFLVTL RLHKATLRHY MALPEQSWHS QLLEFLDVLD DPVLGYLPPT VITILHTHLF SCS VDYRPL YLPVRVLITA ETFTLSSNII MDTSTFLLRF ILDDSALYLS DKCEVETLDL RRDYVCVLDV DLLELVIKTW KGST EGKLS QPLFELRCSN NVVHVHSCAD SCALLVNLLQ YVMSTGDLHP PPRPPSPTEI AGQKLSESPA SLPSCPPVET ALINQ RDLA DALLDTERSL RELAQPSGGH LPQASPISVY LFPGERSGAP PPSPPVGGPA GSLGSCSEEK EDEREEEGDG DTLDSD EFC ILDAPGLGIP PRDGEPVVTQ LHPGPIVVRD GYFSRPIGST DLLRAPAHFP VPSTRVVLRE VSLVWHLYGG RDFGPHP GH RARTGLSGPR SSPSRCSGPN RPQNSWRTQG GSGRQHHVLM EIQLSKVSFQ HEVYPAEPAT GPAAPSQELE ERPLSRQV F IVQELEVRDR LASSQINKFL YLHTSERMPR RAHSNMLTIK ALHVAPTTNL GGPECCLRVS LMPLRLNVDQ DALFFLKDF FTSLVAGINP VVPGETSAEA RPETRAQPSS PLEGQAEGVE TTGSQEAPGG GHSPSPPDQQ PIYFREFRFT SEVPIWLDYH GKHVTMDQV GTFAGLLIGL AQLNCSELKL KRLCCRHGLL GVDKVLGYAL NEWLQDIRKN QLPGLLGGVG PMHSVVQLFQ G FRDLLWLP IEQYRKDGRL MRGLQRGAAS FGSSTASAAL ELSNRLVQAI QATAETVYDI LSPAAPVSRS LQDKRSARRL RR GQQPADL REGVAKAYDT VREGILDTAQ TICDVASRGH EQKGLTGAVG GVIRQLPPTV VKPLILATEA TSSLLGGMRN QIV PDAHKD HALKWRSDSA QD UniProtKB: Autophagy-related protein 2 homolog A |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 61.65 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)