+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3599 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Snake Adenovirus type 1 | |||||||||
 Map data Map data | Snake Adenovirus type 1 | |||||||||
 Sample Sample | Snake Adenovirus type 1 != Snake adenovirus 1 Snake Adenovirus type 1
| |||||||||
| Biological species |  Snake adenovirus 1 Snake adenovirus 1 | |||||||||
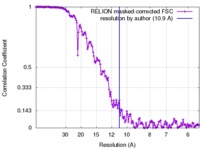
| Method | single particle reconstruction / cryo EM / Resolution: 10.9 Å | |||||||||
 Authors Authors | San Martin C / Menendez-Conejero R | |||||||||
 Citation Citation |  Journal: Structure / Year: 2017 Journal: Structure / Year: 2017Title: Structure of a Reptilian Adenovirus Reveals a Phage Tailspike Fold Stabilizing a Vertebrate Virus Capsid. Authors: Rosa Menéndez-Conejero / Thanh H Nguyen / Abhimanyu K Singh / Gabriela N Condezo / Rachel E Marschang / Mark J van Raaij / Carmen San Martín /    Abstract: Although non-human adenoviruses (AdVs) might offer solutions to problems posed by human AdVs as therapeutic vectors, little is known about their basic biology. In particular, there are no structural ...Although non-human adenoviruses (AdVs) might offer solutions to problems posed by human AdVs as therapeutic vectors, little is known about their basic biology. In particular, there are no structural studies on the complete virion of any AdV with a non-mammalian host. We combine mass spectrometry, cryo-electron microscopy, and protein crystallography to characterize the composition and structure of a snake AdV (SnAdV-1, Atadenovirus genus). SnAdV-1 particles contain the genus-specific proteins LH3, p32k, and LH2, a previously unrecognized structural component. Remarkably, the cementing protein LH3 has a trimeric β helix fold typical of bacteriophage host attachment proteins. The organization of minor coat proteins differs from that in human AdVs, correlating with higher thermostability in SnAdV-1. These findings add a new piece to the intriguing puzzle of virus evolution, hint at the use of cell entry pathways different from those in human AdVs, and will help development of new, thermostable SnAdV-1-based vectors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3599.map.gz emd_3599.map.gz | 232.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3599-v30.xml emd-3599-v30.xml emd-3599.xml emd-3599.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3599_fsc.xml emd_3599_fsc.xml | 14.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_3599.png emd_3599.png | 156.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3599 http://ftp.pdbj.org/pub/emdb/structures/EMD-3599 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3599 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3599 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3599.map.gz / Format: CCP4 / Size: 259.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3599.map.gz / Format: CCP4 / Size: 259.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Snake Adenovirus type 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.75 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Snake Adenovirus type 1
| Entire | Name: Snake Adenovirus type 1 |
|---|---|
| Components |
|
-Supramolecule #1: Snake adenovirus 1
| Supramolecule | Name: Snake adenovirus 1 / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 189830 / Sci species name: Snake adenovirus 1 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Virus shell | Shell ID: 1 / Diameter: 940.0 Å / T number (triangulation number): 25 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Grid | Model: quantifoil / Material: COPPER/RHODIUM / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM CPC |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: KODAK SO-163 FILM / Average electron dose: 12.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 50000 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)