[English] 日本語
 Yorodumi
Yorodumi- EMDB-5538: CryoEM Structure of human defensin 5 bound to a neutralization se... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5538 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM Structure of human defensin 5 bound to a neutralization sensitive adenovirus chimera | |||||||||
 Map data Map data | 3D reconstruction of defensin complexed with a neutralization sensitive adenovirus chimera | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | adenovirus / human defensin 5 / neutralization / cryoEM / MDFF / innate immunity | |||||||||
| Biological species |   Human adenovirus 5 Human adenovirus 5 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 9.6 Å | |||||||||
 Authors Authors | Flatt JW / Smith JG / Nemerow GR / Stewart PL | |||||||||
 Citation Citation |  Journal: PLoS One / Year: 2013 Journal: PLoS One / Year: 2013Title: An intrinsically disordered region of the adenovirus capsid is implicated in neutralization by human alpha defensin 5. Authors: Justin W Flatt / Robert Kim / Jason G Smith / Glen R Nemerow / Phoebe L Stewart /  Abstract: Human α-defensins are proteins of the innate immune system that suppress viral and bacterial infections by multiple mechanisms including membrane disruption. For viruses that lack envelopes, such as ...Human α-defensins are proteins of the innate immune system that suppress viral and bacterial infections by multiple mechanisms including membrane disruption. For viruses that lack envelopes, such as human adenovirus (HAdV), other, less well defined, mechanisms must be involved. A previous structural study on the interaction of an α-defensin, human α-defensin 5 (HD5), with HAdV led to a proposed mechanism in which HD5 stabilizes the vertex region of the capsid and blocks uncoating steps required for infectivity. Studies with virus chimeras comprised of capsid proteins from sensitive and resistant serotypes supported this model. To further characterize the critical binding site, we determined subnanometer resolution cryo-electron microscopy (cryoEM) structures of HD5 complexed with both neutralization-sensitive and -resistant HAdV chimeras. Models were built for the vertex regions of these chimeras with monomeric and dimeric forms of HD5 in various initial orientations. CryoEM guided molecular dynamics flexible fitting (MDFF) was used to restrain the majority of the vertex model in well-defined cryoEM density. The RGD-containing penton base loops of both the sensitive and resistant virus chimeras are predicted to be intrinsically disordered, and little cryoEM density is observed for them. In simulations these loops from the sensitive virus chimera, interact with HD5, bridge the penton base and fiber proteins, and provides significant stabilization with a three-fold increase in the intermolecular nonbonded interactions of the vertex complex. In the case of the resistant virus chimera, simulations revealed fewer bridging interactions and reduced stabilization by HD5. This study implicates a key dynamic region in mediating a stabilizing interaction between a viral capsid and a protein of the innate immune system with potent anti-viral activity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5538.map.gz emd_5538.map.gz | 1.3 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5538-v30.xml emd-5538-v30.xml emd-5538.xml emd-5538.xml | 10.5 KB 10.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5538.jpg emd_5538.jpg | 563.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5538 http://ftp.pdbj.org/pub/emdb/structures/EMD-5538 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5538 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5538 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5538.map.gz / Format: CCP4 / Size: 3.2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5538.map.gz / Format: CCP4 / Size: 3.2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
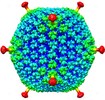
| Annotation | 3D reconstruction of defensin complexed with a neutralization sensitive adenovirus chimera | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
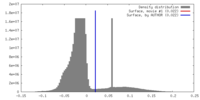
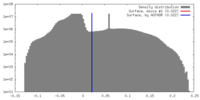
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of an adenovirus chimera Ad5.F35 with human alpha defensin 5
| Entire | Name: Complex of an adenovirus chimera Ad5.F35 with human alpha defensin 5 |
|---|---|
| Components |
|
-Supramolecule #1000: Complex of an adenovirus chimera Ad5.F35 with human alpha defensin 5
| Supramolecule | Name: Complex of an adenovirus chimera Ad5.F35 with human alpha defensin 5 type: sample / ID: 1000 Oligomeric state: ~3000 molecules bind each adenovirus particle Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 150 MDa |
-Supramolecule #1: Human adenovirus 5
| Supramolecule | Name: Human adenovirus 5 / type: virus / ID: 1 / Name.synonym: Human adenovirus type 5 with short fiber / NCBI-ID: 28285 / Sci species name: Human adenovirus 5 / Database: NCBI / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No / Syn species name: Human adenovirus type 5 with short fiber |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
| Molecular weight | Theoretical: 150 MDa |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 1170 Å / T number (triangulation number): 25 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.16 mg/mL |
|---|---|
| Buffer | pH: 7.6 / Details: 50mM Tris, 150mM NaCl, 2mM CaCl2, 2mM MgCl2 |
| Grid | Details: Quantifoil R2/4 holey carbon grids, glow discharged |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 30 % / Chamber temperature: 90 K / Instrument: HOMEMADE PLUNGER / Method: Blot for 4 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 300,000 times magnification Legacy - Electron beam tilt params: 0 |
| Details | Low dose |
| Date | Jul 12, 2010 |
| Image recording | Category: FILM / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Scanner: OTHER / Digitization - Sampling interval: 15 µm / Number real images: 3515 / Average electron dose: 20 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 400000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.26 mm / Nominal defocus max: 2.1 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 310000 |
| Sample stage | Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The particles were selected with an in-house script and processed using Frealign |
|---|---|
| CTF correction | Details: Each particle |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 9.6 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Frealign / Number images used: 1014 |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)