[English] 日本語
 Yorodumi
Yorodumi- EMDB-5511: CryoEM Visualization of an Adenovirus Capsid-Incorporated HIV Antigen -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5511 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM Visualization of an Adenovirus Capsid-Incorporated HIV Antigen | |||||||||
 Map data Map data | 3D reconstruction of an adenovirus vector engineered for a vaccine application | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cryoEM / viral vaccine vector / adenovirus / HIV / gp41 / MPER | |||||||||
| Biological species |   Human adenovirus 5 Human adenovirus 5 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.7 Å | |||||||||
 Authors Authors | Flatt JW / Fox TL / Makarova N / Blackwell JL / Dmitriev IP / Kashentseva EA / Curiel DT / Stewart PL | |||||||||
 Citation Citation |  Journal: PLoS One / Year: 2012 Journal: PLoS One / Year: 2012Title: CryoEM visualization of an adenovirus capsid-incorporated HIV antigen. Authors: Justin W Flatt / Tara L Fox / Natalia Makarova / Jerry L Blackwell / Igor P Dmitriev / Elena A Kashentseva / David T Curiel / Phoebe L Stewart /  Abstract: Adenoviral (Ad) vectors show promise as platforms for vaccine applications against infectious diseases including HIV. However, the requirements for eliciting protective neutralizing antibody and ...Adenoviral (Ad) vectors show promise as platforms for vaccine applications against infectious diseases including HIV. However, the requirements for eliciting protective neutralizing antibody and cellular immune responses against HIV remain a major challenge. In a novel approach to generate 2F5- and 4E10-like antibodies, we engineered an Ad vector with the HIV membrane proximal ectodomain region (MPER) epitope displayed on the hypervariable region 2 (HVR2) of the viral hexon capsid, instead of expressed as a transgene. The structure and flexibility of MPER epitopes, and the structural context of these epitopes within viral vectors, play important roles in the induced host immune responses. In this regard, understanding the critical factors for epitope presentation would facilitate optimization strategies for developing viral vaccine vectors. Therefore we undertook a cryoEM structural study of this Ad vector, which was previously shown to elicit MPER-specific humoral immune responses. A subnanometer resolution cryoEM structure was analyzed with guided molecular dynamics simulations. Due to the arrangement of hexons within the Ad capsid, there are twelve unique environments for the inserted peptide that lead to a variety of conformations for MPER, including individual α-helices, interacting α-helices, and partially extended forms. This finding is consistent with the known conformational flexibility of MPER. The presence of an extended form, or an induced extended form, is supported by interaction of this vector with the human HIV monoclonal antibody 2F5, which recognizes 14 extended amino acids within MPER. These results demonstrate that the Ad capsid influences epitope structure, flexibility and accessibility, all of which affect the host immune response. In summary, this cryoEM structural study provided a means to visualize an epitope presented on an engineered viral vector and suggested modifications for the next generation of Ad vectors with capsid-incorporated HIV epitopes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5511.map.gz emd_5511.map.gz | 91.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5511-v30.xml emd-5511-v30.xml emd-5511.xml emd-5511.xml | 10.6 KB 10.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5511.jpg emd_5511.jpg | 233.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5511 http://ftp.pdbj.org/pub/emdb/structures/EMD-5511 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5511 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5511 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5511.map.gz / Format: CCP4 / Size: 976.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5511.map.gz / Format: CCP4 / Size: 976.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D reconstruction of an adenovirus vector engineered for a vaccine application | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.25 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
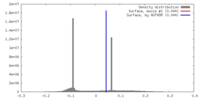
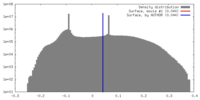
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ad vector with a capsid-incorporated HIV epitope based on the mem...
| Entire | Name: Ad vector with a capsid-incorporated HIV epitope based on the membrane proximal ectodomain region (MPER) of gp41 |
|---|---|
| Components |
|
-Supramolecule #1000: Ad vector with a capsid-incorporated HIV epitope based on the mem...
| Supramolecule | Name: Ad vector with a capsid-incorporated HIV epitope based on the membrane proximal ectodomain region (MPER) of gp41 type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 150 MDa |
-Supramolecule #1: Human adenovirus 5
| Supramolecule | Name: Human adenovirus 5 / type: virus / ID: 1 Details: recombinant type 5 adenovirus vector, Ad-HVR2-GP41-L15, generated with a 24 amino acid MPER insertion within hypervariable region 2 (HVR2) of the hexon capsid protein NCBI-ID: 28285 / Sci species name: Human adenovirus 5 / Database: NCBI / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
| Molecular weight | Theoretical: 150 MDa |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 1170 Å / T number (triangulation number): 25 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.6 / Details: 50 mM Tris, 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2 |
| Grid | Details: Quantifoil R2/4 holey carbon grids, glow discharged |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 30 % / Chamber temperature: 90 K / Instrument: HOMEMADE PLUNGER / Method: Blot for 4 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Temperature | Average: 90 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 300,000 times magnification Legacy - Electron beam tilt params: 0 |
| Date | Jun 1, 2011 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Sampling interval: 15 µm / Number real images: 5025 / Average electron dose: 20 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 400000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.26 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 310000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The particles were selected with in-house script and processed using Frealign |
|---|---|
| CTF correction | Details: Each particle |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 8.7 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Frealign / Number images used: 1306 |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)