[English] 日本語
 Yorodumi
Yorodumi- EMDB-34463: Cryo-EM Structure of the KBTBD2-CRL3~N8(removed)-CSN(no CSN3/8) c... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of the KBTBD2-CRL3~N8(removed)-CSN(no CSN3/8) complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ligase / complex | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.15 Å | |||||||||
 Authors Authors | Hu Y / Mao Q / Chen Z / Sun L | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3. Authors: Yuxia Hu / Zhao Zhang / Qiyu Mao / Xiang Zhang / Aihua Hao / Yu Xun / Yeda Wang / Lin Han / Wuqiang Zhan / Qianying Liu / Yue Yin / Chao Peng / Eva Marie Y Moresco / Zhenguo Chen / Bruce Beutler / Lei Sun /   Abstract: Phosphatidylinositol 3-kinase α, a heterodimer of catalytic p110α and one of five regulatory subunits, mediates insulin- and insulin like growth factor-signaling and, frequently, oncogenesis. ...Phosphatidylinositol 3-kinase α, a heterodimer of catalytic p110α and one of five regulatory subunits, mediates insulin- and insulin like growth factor-signaling and, frequently, oncogenesis. Cellular levels of the regulatory p85α subunit are tightly controlled by regulated proteasomal degradation. In adipose tissue and growth plates, failure of K48-linked p85α ubiquitination causes diabetes, lipodystrophy and dwarfism in mice, as in humans with SHORT syndrome. Here we elucidated the structures of the key ubiquitin ligase complexes regulating p85α availability. Specificity is provided by the substrate receptor KBTBD2, which recruits p85α to the cullin3-RING E3 ubiquitin ligase (CRL3). CRL3 forms multimers, which disassemble into dimers upon substrate binding (CRL3-p85α) and/or neddylation by the activator NEDD8 (CRL3~N8), leading to p85α ubiquitination and degradation. Deactivation involves dissociation of NEDD8 mediated by the COP9 signalosome and displacement of KBTBD2 by the inhibitor CAND1. The hereby identified structural basis of p85α regulation opens the way to better understanding disturbances of glucose regulation, growth and cancer. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34463.map.gz emd_34463.map.gz | 35.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34463-v30.xml emd-34463-v30.xml emd-34463.xml emd-34463.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34463.png emd_34463.png | 79.3 KB | ||
| Filedesc metadata |  emd-34463.cif.gz emd-34463.cif.gz | 4.1 KB | ||
| Others |  emd_34463_half_map_1.map.gz emd_34463_half_map_1.map.gz emd_34463_half_map_2.map.gz emd_34463_half_map_2.map.gz | 29.6 MB 29.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34463 http://ftp.pdbj.org/pub/emdb/structures/EMD-34463 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34463 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34463 | HTTPS FTP |
-Validation report
| Summary document |  emd_34463_validation.pdf.gz emd_34463_validation.pdf.gz | 686.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34463_full_validation.pdf.gz emd_34463_full_validation.pdf.gz | 686.2 KB | Display | |
| Data in XML |  emd_34463_validation.xml.gz emd_34463_validation.xml.gz | 10.9 KB | Display | |
| Data in CIF |  emd_34463_validation.cif.gz emd_34463_validation.cif.gz | 12.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34463 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34463 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34463 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34463 | HTTPS FTP |
-Related structure data
| Related structure data |  8gq6C  8h33C  8h34C  8h35C  8h36C  8h37C  8h38C  8h3aC  8h3fC  8h3qC  8h3rC C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34463.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34463.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.088 Å | ||||||||||||||||||||||||||||||||||||
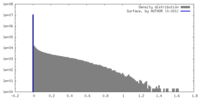
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34463_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
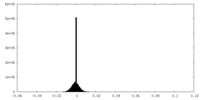
| Density Histograms |
-Half map: #2
| File | emd_34463_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
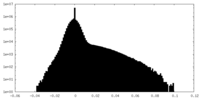
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM Structure of the KBTBD2-CRL3~N8-CSN(no CSN3/8) complex
| Entire | Name: Cryo-EM Structure of the KBTBD2-CRL3~N8-CSN(no CSN3/8) complex |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM Structure of the KBTBD2-CRL3~N8-CSN(no CSN3/8) complex
| Supramolecule | Name: Cryo-EM Structure of the KBTBD2-CRL3~N8-CSN(no CSN3/8) complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#11 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: CSN
| Supramolecule | Name: CSN / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#8 |
|---|
-Supramolecule #3: KBTBD2,CRL3
| Supramolecule | Name: KBTBD2,CRL3 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #9-#11 |
|---|
-Supramolecule #4: N8
| Supramolecule | Name: N8 / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #12 |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: Details: 6r7f |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 7.15 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 63653 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)