+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3329 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo electron microscopy of a complex of Tor and Lst8 | |||||||||
 Map data Map data | volume for dimer of Tor/Lst8 heterodimers | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cryo-EM / Tor / Lst8 / mTOR / kinase / PIKK / S/T protein kinase / TORC1 / mTORC1 | |||||||||
| Function / homology | TORC1 complex / : / :  Function and homology information Function and homology information | |||||||||
| Biological species |  Kluyveromyces marxianus (yeast) Kluyveromyces marxianus (yeast) | |||||||||
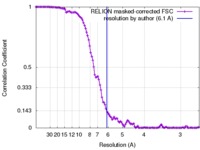
| Method | single particle reconstruction / cryo EM / Resolution: 6.1 Å | |||||||||
 Authors Authors | Baretic D / Berndt A / Ohashi Y / Johnson CM / Williams RL | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2016 Journal: Nat Commun / Year: 2016Title: Tor forms a dimer through an N-terminal helical solenoid with a complex topology. Authors: Domagoj Baretić / Alex Berndt / Yohei Ohashi / Christopher M Johnson / Roger L Williams /  Abstract: The target of rapamycin (Tor) is a Ser/Thr protein kinase that regulates a range of anabolic and catabolic processes. Tor is present in two complexes, TORC1 and TORC2, in which the Tor-Lst8 ...The target of rapamycin (Tor) is a Ser/Thr protein kinase that regulates a range of anabolic and catabolic processes. Tor is present in two complexes, TORC1 and TORC2, in which the Tor-Lst8 heterodimer forms a common sub-complex. We have determined the cryo-electron microscopy (EM) structure of Tor bound to Lst8. Two Tor-Lst8 heterodimers assemble further into a dyad-symmetry dimer mediated by Tor-Tor interactions. The first 1,300 residues of Tor form a HEAT repeat-containing α-solenoid with four distinct segments: a highly curved 800-residue N-terminal 'spiral', followed by a 400-residue low-curvature 'bridge' and an extended 'railing' running along the bridge leading to the 'cap' that links to FAT region. This complex topology was verified by domain insertions and offers a new interpretation of the mTORC1 structure. The spiral of one TOR interacts with the bridge of another, which together form a joint platform for the Regulatory Associated Protein of TOR (RAPTOR) regulatory subunit. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3329.map.gz emd_3329.map.gz | 116.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3329-v30.xml emd-3329-v30.xml emd-3329.xml emd-3329.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3329_fsc.xml emd_3329_fsc.xml | 11 KB | Display |  FSC data file FSC data file |
| Images |  EMD-3329_image.png EMD-3329_image.png | 69 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3329 http://ftp.pdbj.org/pub/emdb/structures/EMD-3329 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3329 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3329 | HTTPS FTP |
-Validation report
| Summary document |  emd_3329_validation.pdf.gz emd_3329_validation.pdf.gz | 294 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3329_full_validation.pdf.gz emd_3329_full_validation.pdf.gz | 293.2 KB | Display | |
| Data in XML |  emd_3329_validation.xml.gz emd_3329_validation.xml.gz | 11.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3329 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3329 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3329 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3329 | HTTPS FTP |
-Related structure data
| Related structure data |  5fvmMC  3334C  3335C  3336C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3329.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3329.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | volume for dimer of Tor/Lst8 heterodimers | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.33 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of Tor1 with Lst8
| Entire | Name: Complex of Tor1 with Lst8 |
|---|---|
| Components |
|
-Supramolecule #1000: Complex of Tor1 with Lst8
| Supramolecule | Name: Complex of Tor1 with Lst8 / type: sample / ID: 1000 / Oligomeric state: Dimer of Tor1/Lst8 heterodimers / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 622 KDa |
-Macromolecule #1: Target of rapamycin (Tor)
| Macromolecule | Name: Target of rapamycin (Tor) / type: protein_or_peptide / ID: 1 / Name.synonym: Tor1 / Number of copies: 2 / Oligomeric state: dimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Kluyveromyces marxianus (yeast) Kluyveromyces marxianus (yeast) |
| Molecular weight | Theoretical: 277 KDa |
| Recombinant expression | Organism:  Kluyveromyces marxianus (yeast) Kluyveromyces marxianus (yeast) |
| Sequence | GO: TORC1 complex |
-Macromolecule #2: Lst8
| Macromolecule | Name: Lst8 / type: protein_or_peptide / ID: 2 / Name.synonym: Lethal with SEC13 protein 8 / Number of copies: 2 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Kluyveromyces marxianus (yeast) Kluyveromyces marxianus (yeast) |
| Molecular weight | Theoretical: 34 KDa |
| Recombinant expression | Organism:  Kluyveromyces marxianus (yeast) Kluyveromyces marxianus (yeast) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 50 mM Hepes pH 7.4 (23 deg C), 75 mM KCl, 250 mM NaCl, 0.3 % (v/v) CHAPS, 1 mM TCEP |
| Grid | Details: Quantifoil Au R 0.6/1.0 or Au R 1.2/1.3, 300 mesh grids, blotted for 11-13 s at 4 deg C |
| Vitrification | Cryogen name: ETHANE / Instrument: OTHER / Method: 11-13 s at 4 deg C |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Date | May 16, 2015 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 2332 / Average electron dose: 40 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 105263 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 47000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | An initial model for the FATKIN was derived from 4JSV. A poly-ALA model was built into the remaining density (about half of the total). The full model was refined by Phenix real space, global minimization, 2-fold NCS, one B-factor per residue, secondary structure rstraints, ramachandran restraints |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: correlation |
| Output model |  PDB-5fvm: |
-Atomic model buiding 2
| Initial model | PDB ID: Chain - #0 - Chain ID: C / Chain - #1 - Chain ID: D |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | An initial model for the Lst8 was derived from 4JSV. A poly-ALA model was adjusted to the density. The full model was refined by Phenix real space, global minimization, 2-fold NCS, one B-factor per residue, secondary structure rstraints, ramachandran restraints |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: correlation |
| Output model |  PDB-5fvm: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)