+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3284 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of BK polyomavirus VP1 virus-like particle | ||||||||||||
 Map data Map data | Reconstruction of BK VP1 VLP (sharpened/masked) | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | BKPyV / BK / polyomavirus / virus-like particle / VLP | ||||||||||||
| Biological species | BK polyomavirus VP1 VLP | ||||||||||||
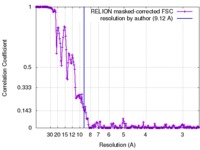
| Method | single particle reconstruction / cryo EM / Resolution: 9.12 Å | ||||||||||||
 Authors Authors | Hurdiss DL / Morgan EL / Thompson RF / Prescott EL / Panou MM / Macdonald A / Ranson NA | ||||||||||||
 Citation Citation |  Journal: Structure / Year: 2016 Journal: Structure / Year: 2016Title: New Structural Insights into the Genome and Minor Capsid Proteins of BK Polyomavirus using Cryo-Electron Microscopy. Authors: Daniel L Hurdiss / Ethan L Morgan / Rebecca F Thompson / Emma L Prescott / Margarita M Panou / Andrew Macdonald / Neil A Ranson /  Abstract: BK polyomavirus is the causative agent of several diseases in transplant patients and the immunosuppressed. In order to better understand the structure and life cycle of BK, we produced infectious ...BK polyomavirus is the causative agent of several diseases in transplant patients and the immunosuppressed. In order to better understand the structure and life cycle of BK, we produced infectious virions and VP1-only virus-like particles in cell culture, and determined their three-dimensional structures using cryo-electron microscopy (EM) and single-particle image processing. The resulting 7.6-Å resolution structure of BK and 9.1-Å resolution of the virus-like particles are the highest-resolution cryo-EM structures of any polyomavirus. These structures confirm that the architecture of the major structural protein components of these human polyomaviruses are similar to previous structures from other hosts, but give new insight into the location and role of the enigmatic minor structural proteins, VP2 and VP3. We also observe two shells of electron density, which we attribute to a structurally ordered part of the viral genome, and discrete contacts between this density and both VP1 and the minor capsid proteins. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3284.map.gz emd_3284.map.gz | 104.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3284-v30.xml emd-3284-v30.xml emd-3284.xml emd-3284.xml | 8.8 KB 8.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3284_fsc.xml emd_3284_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_3284.tif emd_3284.tif | 2.2 MB | ||
| Others |  emd_3284_additional_1.map.gz emd_3284_additional_1.map.gz | 374.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3284 http://ftp.pdbj.org/pub/emdb/structures/EMD-3284 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3284 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3284 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3284.map.gz / Format: CCP4 / Size: 465.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3284.map.gz / Format: CCP4 / Size: 465.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of BK VP1 VLP (sharpened/masked) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Supplemental map: emd 3284 additional 1.map
| File | emd_3284_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BK polyomavirus VP1 VLP
| Entire | Name: BK polyomavirus VP1 VLP |
|---|---|
| Components |
|
-Supramolecule #1000: BK polyomavirus VP1 VLP
| Supramolecule | Name: BK polyomavirus VP1 VLP / type: sample / ID: 1000 / Oligomeric state: Icosohedral / Number unique components: 1 |
|---|
-Supramolecule #1: BK polyomavirus VP1 VLP
| Supramolecule | Name: BK polyomavirus VP1 VLP / type: virus / ID: 1 / Sci species name: BK polyomavirus VP1 VLP / Sci species strain: BKV-Ia / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
| Host system | Recombinant cell: HEK293TT / Recombinant plasmid: pIaw |
| Virus shell | Shell ID: 1 / Diameter: 498 Å |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 / Details: 10mM HEPES pH 7.9, 50mM CaCl2, 1mM MgCl2, 5mM KCl |
|---|---|
| Grid | Details: Quantifoil R2/1 EM grids |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV / Method: 6.5 seconds blot before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Date | May 13, 2015 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number real images: 170 / Average electron dose: 40 e/Å2 Details: 4 e-/A2/s, a 4 frames per second frame rate, and a 10 s exposure |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 6.136 µm / Nominal defocus min: 0.526 µm / Nominal magnification: 19000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)