+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32190 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Coxsackievirus B3 full particle at pH7.4 (VP3-234N) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CVB3 / VP3-234N / full particle / VIRUS | |||||||||
| Biological species |   Coxsackievirus B3 Coxsackievirus B3 | |||||||||
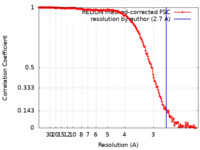
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Wang QL / Liu CC | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains. Authors: Qingling Wang / Qian Yang / Congcong Liu / Guoqing Wang / Hao Song / Guijun Shang / Ruchao Peng / Xiao Qu / Sheng Liu / Yingzi Cui / Peiyi Wang / Wenbo Xu / Xin Zhao / Jianxun Qi / Mengsu Yang / George F Gao /  Abstract: Receptor usage defines cell tropism and contributes to cell entry and infection. Coxsackievirus B (CVB) engages coxsackievirus and adenovirus receptor (CAR), and selectively utilizes the decay- ...Receptor usage defines cell tropism and contributes to cell entry and infection. Coxsackievirus B (CVB) engages coxsackievirus and adenovirus receptor (CAR), and selectively utilizes the decay-accelerating factor (DAF; CD55) to infect cells. However, the differential receptor usage mechanism for CVB remains elusive. This study identified VP3-234 residues (234Q/N/V/D/E) as critical population selection determinants during CVB3 virus evolution, contributing to diverse binding affinities to CD55. Cryoelectron microscopy (cryo-EM) structures of CD55-binding/nonbinding isolates and their complexes with CD55 or CAR were obtained under both neutral and acidic conditions, and the molecular mechanism of VP3-234 residues determining CD55 affinity/specificity for naturally occurring CVB3 strains was elucidated. Structural and biochemical studies in vitro revealed the dynamic entry process of CVB3 and the function of the uncoating receptor CAR with different pH preferences. This work provides detailed insight into the molecular mechanism of CVB infection and contributes to an in-depth understanding of enterovirus attachment receptor usage. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32190.map.gz emd_32190.map.gz | 59.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32190-v30.xml emd-32190-v30.xml emd-32190.xml emd-32190.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32190_fsc.xml emd_32190_fsc.xml | 15.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_32190.png emd_32190.png | 108.6 KB | ||
| Masks |  emd_32190_msk_1.map emd_32190_msk_1.map | 307.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-32190.cif.gz emd-32190.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32190 http://ftp.pdbj.org/pub/emdb/structures/EMD-32190 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32190 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32190 | HTTPS FTP |
-Validation report
| Summary document |  emd_32190_validation.pdf.gz emd_32190_validation.pdf.gz | 641.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32190_full_validation.pdf.gz emd_32190_full_validation.pdf.gz | 641.5 KB | Display | |
| Data in XML |  emd_32190_validation.xml.gz emd_32190_validation.xml.gz | 14.6 KB | Display | |
| Data in CIF |  emd_32190_validation.cif.gz emd_32190_validation.cif.gz | 19.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32190 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32190 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32190 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32190 | HTTPS FTP |
-Related structure data
| Related structure data |  7vy0MC  7vxhC  7vxzC  7vy5C  7vy6C  7vykC  7vylC  7vymC  7w14C  7w17C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32190.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32190.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_32190_msk_1.map emd_32190_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Coxsackievirus B3
| Entire | Name:   Coxsackievirus B3 Coxsackievirus B3 |
|---|---|
| Components |
|
-Supramolecule #1: Coxsackievirus B3
| Supramolecule | Name: Coxsackievirus B3 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 / NCBI-ID: 12072 / Sci species name: Coxsackievirus B3 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Coxsackievirus B3 Coxsackievirus B3 |
| Molecular weight | Theoretical: 31.703477 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPVEDVITAA IGRVADTVGT GPTNSEAIPA LTAAETGHTS QVVPGDTMQT RHVKNYHSRS ESTVENFLCR SACVYFTEYK NSGSKRYAE WVVTTRQAAQ LRRKLEFFTY IRFDLELTFV ITSTQQPSTT QNQDAQILTH QIMYVPPGGP VPDKVDSYVW Q TSTNPSVF ...String: GPVEDVITAA IGRVADTVGT GPTNSEAIPA LTAAETGHTS QVVPGDTMQT RHVKNYHSRS ESTVENFLCR SACVYFTEYK NSGSKRYAE WVVTTRQAAQ LRRKLEFFTY IRFDLELTFV ITSTQQPSTT QNQDAQILTH QIMYVPPGGP VPDKVDSYVW Q TSTNPSVF WTEGNAPPRM SIPFLSIGNA YSNFYDGWSE FSRNGVYGIN TLNNMGTLYA RHVNTGSTGP IKSTIRIYFK PK HVKAWIP RPPRLCQYEK AKNVNFQPSG VTTTRQSITT MTNTGAF |
-Macromolecule #2: Capsid protein VP2
| Macromolecule | Name: Capsid protein VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Coxsackievirus B3 Coxsackievirus B3 |
| Molecular weight | Theoretical: 28.862561 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SPTVEECGYS DRVRSITLGN STITTQECAN VVVGYGVWPD YLKDSEATAE DQPTQPDVAT CRFYTLDSVQ WQKTSPGWWW KLPDALSNL GLFGQNMQYH YLGRTGYTIH VQCNASKFHQ GCLLVVCVPE AEMGCATLDN TPSSAELLGG DAAKEFAGEP I ASGSNKLV ...String: SPTVEECGYS DRVRSITLGN STITTQECAN VVVGYGVWPD YLKDSEATAE DQPTQPDVAT CRFYTLDSVQ WQKTSPGWWW KLPDALSNL GLFGQNMQYH YLGRTGYTIH VQCNASKFHQ GCLLVVCVPE AEMGCATLDN TPSSAELLGG DAAKEFAGEP I ASGSNKLV QRVVYNAGMG IGVGNLTIFP HQWINLRTNN SATIVMPYTN SVPMDNMFRH NNITLMVIPF VPLDYCPGST TY VPITVTI APMCAEYNGL RLASHQ |
-Macromolecule #3: Capsid protein VP3
| Macromolecule | Name: Capsid protein VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Coxsackievirus B3 Coxsackievirus B3 |
| Molecular weight | Theoretical: 26.128633 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GLPTMNTPGS CQFLTSDDFQ SPSAMPQYDV TPEMKIPGEV KNLMEIAEVD SVVPVQNVGE KVNSMEAYQI PVRSNEGSGT QVFGFPLQP GYSSVFSRTL LGEILNYYTH WSGSIKLTFM FCGSAMATGK FLLAYSPPGA GAPTKRVDAM LGTHVVWDVG L QSSCVLCI ...String: GLPTMNTPGS CQFLTSDDFQ SPSAMPQYDV TPEMKIPGEV KNLMEIAEVD SVVPVQNVGE KVNSMEAYQI PVRSNEGSGT QVFGFPLQP GYSSVFSRTL LGEILNYYTH WSGSIKLTFM FCGSAMATGK FLLAYSPPGA GAPTKRVDAM LGTHVVWDVG L QSSCVLCI PWISQTHYRY VASDEYTAGG FITCWYQTNI VVPADAQSSC YIMCFVSACN DFSVRLLKDT PFISQNSFFQ |
-Macromolecule #4: Capsid protein VP4
| Macromolecule | Name: Capsid protein VP4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Coxsackievirus B3 Coxsackievirus B3 |
| Molecular weight | Theoretical: 7.437209 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGAQVSTQKT GAHETGLNAS GNSIIHYTNI NYYKDAASNS ATRQDFAQDP GKFTEPVKDI MIKSLPALN |
-Macromolecule #5: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 5 / Number of copies: 1 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Support film - Material: CARBON / Support film - topology: CONTINUOUS |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average exposure time: 1.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 5.0 µm / Calibrated defocus min: 1.8 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT | ||||||||||
| Output model |  PDB-7vy0: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)