[English] 日本語
 Yorodumi
Yorodumi- EMDB-32207: Coxsackievirus B3 at pH7.4 (VP3-234Q) incubation with coxsackievi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32207 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Coxsackievirus B3 at pH7.4 (VP3-234Q) incubation with coxsackievirus and adenovirus receptor for 10min | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CVB3 / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationAV node cell-bundle of His cell adhesion involved in cell communication / cell adhesive protein binding involved in AV node cell-bundle of His cell communication / symbiont-mediated perturbation of host transcription / AV node cell to bundle of His cell communication / homotypic cell-cell adhesion / epithelial structure maintenance / regulation of AV node cell action potential / gamma-delta T cell activation / apicolateral plasma membrane / germ cell migration ...AV node cell-bundle of His cell adhesion involved in cell communication / cell adhesive protein binding involved in AV node cell-bundle of His cell communication / symbiont-mediated perturbation of host transcription / AV node cell to bundle of His cell communication / homotypic cell-cell adhesion / epithelial structure maintenance / regulation of AV node cell action potential / gamma-delta T cell activation / apicolateral plasma membrane / germ cell migration / connexin binding / transepithelial transport / cell-cell junction organization / cardiac muscle cell development / heterophilic cell-cell adhesion / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / intercalated disc / bicellular tight junction / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / neutrophil chemotaxis / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / cell adhesion molecule binding / acrosomal vesicle / Cell surface interactions at the vascular wall / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / PDZ domain binding / filopodium / adherens junction / mitochondrion organization / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / neuromuscular junction / T=pseudo3 icosahedral viral capsid / beta-catenin binding / host cell cytoplasmic vesicle membrane / integrin binding / Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell / ribonucleoside triphosphate phosphatase activity / cell-cell junction / cell junction / nucleoside-triphosphate phosphatase / heart development / channel activity / growth cone / virus receptor activity / cell body / actin cytoskeleton organization / monoatomic ion transmembrane transport / basolateral plasma membrane / defense response to virus / symbiont-mediated suppression of host NF-kappaB cascade / DNA replication / RNA helicase activity / neuron projection / membrane raft / endocytosis involved in viral entry into host cell / signaling receptor binding / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / protein-containing complex / proteolysis / extracellular space / RNA binding / extracellular region / zinc ion binding / nucleoplasm / ATP binding / identical protein binding / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Coxsackievirus B3 / Coxsackievirus B3 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
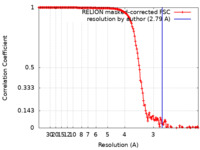
| Method | single particle reconstruction / cryo EM / Resolution: 2.79 Å | |||||||||
 Authors Authors | Wang QL / Liu CC | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains. Authors: Qingling Wang / Qian Yang / Congcong Liu / Guoqing Wang / Hao Song / Guijun Shang / Ruchao Peng / Xiao Qu / Sheng Liu / Yingzi Cui / Peiyi Wang / Wenbo Xu / Xin Zhao / Jianxun Qi / Mengsu Yang / George F Gao /  Abstract: Receptor usage defines cell tropism and contributes to cell entry and infection. Coxsackievirus B (CVB) engages coxsackievirus and adenovirus receptor (CAR), and selectively utilizes the decay- ...Receptor usage defines cell tropism and contributes to cell entry and infection. Coxsackievirus B (CVB) engages coxsackievirus and adenovirus receptor (CAR), and selectively utilizes the decay-accelerating factor (DAF; CD55) to infect cells. However, the differential receptor usage mechanism for CVB remains elusive. This study identified VP3-234 residues (234Q/N/V/D/E) as critical population selection determinants during CVB3 virus evolution, contributing to diverse binding affinities to CD55. Cryoelectron microscopy (cryo-EM) structures of CD55-binding/nonbinding isolates and their complexes with CD55 or CAR were obtained under both neutral and acidic conditions, and the molecular mechanism of VP3-234 residues determining CD55 affinity/specificity for naturally occurring CVB3 strains was elucidated. Structural and biochemical studies in vitro revealed the dynamic entry process of CVB3 and the function of the uncoating receptor CAR with different pH preferences. This work provides detailed insight into the molecular mechanism of CVB infection and contributes to an in-depth understanding of enterovirus attachment receptor usage. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32207.map.gz emd_32207.map.gz | 86.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32207-v30.xml emd-32207-v30.xml emd-32207.xml emd-32207.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32207_fsc.xml emd_32207_fsc.xml | 15.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_32207.png emd_32207.png | 111.1 KB | ||
| Masks |  emd_32207_msk_1.map emd_32207_msk_1.map | 307.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-32207.cif.gz emd-32207.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32207 http://ftp.pdbj.org/pub/emdb/structures/EMD-32207 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32207 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32207 | HTTPS FTP |
-Related structure data
| Related structure data |  7vykMC  7vxhC  7vxzC  7vy0C  7vy5C  7vy6C  7vylC  7vymC  7w14C  7w17C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32207.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32207.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_32207_msk_1.map emd_32207_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Coxsackievirus B3
| Entire | Name:   Coxsackievirus B3 Coxsackievirus B3 |
|---|---|
| Components |
|
-Supramolecule #1: Coxsackievirus B3
| Supramolecule | Name: Coxsackievirus B3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:   Coxsackievirus B3 Coxsackievirus B3 |
-Supramolecule #2: Capsid protein VP1, VP2, VP3, VP4
| Supramolecule | Name: Capsid protein VP1, VP2, VP3, VP4 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: coxsackievirus and adenovirus receptor
| Supramolecule | Name: coxsackievirus and adenovirus receptor / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #5 |
|---|
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Coxsackievirus B3 Coxsackievirus B3 |
| Molecular weight | Theoretical: 30.05366 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RVADTVGTGP TNSEAIPALT AAETGHTSQV VPGDTMQTRH VKNYHSRSES TIENFLCRSA CVYFTEYENS GAKRYAEWVL TPRQAAQLR RKLEFFTYVR FDLELTFVIT STQQPSTTQN QDAQILTHQI MYVPPGGPVP DKVDSYVWQT STNPSVFWTE G NAPPRMSI ...String: RVADTVGTGP TNSEAIPALT AAETGHTSQV VPGDTMQTRH VKNYHSRSES TIENFLCRSA CVYFTEYENS GAKRYAEWVL TPRQAAQLR RKLEFFTYVR FDLELTFVIT STQQPSTTQN QDAQILTHQI MYVPPGGPVP DKVDSYVWQT STNPSVFWTE G NAPPRMSI PFLSIGNAYS NFYDGWSEFS RNGVYGINTL NNMGTLYARH VNAGSTGPIK STIRIYFKPK HVKAWIPRPP RL CQYEKAK NVNFQPSGVT TTRQSITTMT UniProtKB: Genome polyprotein |
-Macromolecule #2: Capsid protein VP2
| Macromolecule | Name: Capsid protein VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Coxsackievirus B3 Coxsackievirus B3 |
| Molecular weight | Theoretical: 28.822475 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SPTVEECGYS DRARSITLGN STITTQECAN VVVGYGVWPD YLKDSEATAE DQPTQPDVAT CRFYTLDSVQ WQKTSPGWWW KLPDALSNL GLFGQNMQYH YLGRTGYTVH VQCNASKFHQ GCLLVVCVPE AEMGCATLDN TPSSAELLGG DSAKEFADKP V ASGSNKLV ...String: SPTVEECGYS DRARSITLGN STITTQECAN VVVGYGVWPD YLKDSEATAE DQPTQPDVAT CRFYTLDSVQ WQKTSPGWWW KLPDALSNL GLFGQNMQYH YLGRTGYTVH VQCNASKFHQ GCLLVVCVPE AEMGCATLDN TPSSAELLGG DSAKEFADKP V ASGSNKLV QRVVYNAGMG VGVGNLTIFP HQWINLRTNN SATIVMPYTN SVPMDNMFRH NNVTLMVIPF VPLDYCPGST TY VPITVTI APMCAEYNGL RLAGHQ UniProtKB: Genome polyprotein |
-Macromolecule #3: Capsid protein VP3
| Macromolecule | Name: Capsid protein VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Coxsackievirus B3 Coxsackievirus B3 |
| Molecular weight | Theoretical: 26.227725 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GLPTMNTPGS CQFLTSDDFQ SPSAMPQYDV TPEMRIPGEV KNLMEIAEVD SVVPVQNVGE KVNSMEAYQI PVRSNEGSGT QVFGFPLQP GYSSVFSRTL LGEILNYYTH WSGSIKLTFM FCGSAMATGK FLLAYSPPGA GAPTKRVDAM LGTHVVWDVG L QSSCVLCI ...String: GLPTMNTPGS CQFLTSDDFQ SPSAMPQYDV TPEMRIPGEV KNLMEIAEVD SVVPVQNVGE KVNSMEAYQI PVRSNEGSGT QVFGFPLQP GYSSVFSRTL LGEILNYYTH WSGSIKLTFM FCGSAMATGK FLLAYSPPGA GAPTKRVDAM LGTHVVWDVG L QSSCVLCI PWISQTHYRY VTSDEYTAGG FITCWYQTNI VVPADAQSSC YIMCFVSACN DFSVRLLKDT PFISQQNFFQ UniProtKB: Genome polyprotein |
-Macromolecule #4: Capsid protein VP4
| Macromolecule | Name: Capsid protein VP4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Coxsackievirus B3 Coxsackievirus B3 |
| Molecular weight | Theoretical: 7.480235 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGAQVSTQKT GAHETGLNAS GNSIIHYTNI NYYKDAASNS ANRQDFTQDP GKFTEPVKDI MIKSLPALN UniProtKB: Genome polyprotein |
-Macromolecule #5: Coxsackievirus and adenovirus receptor
| Macromolecule | Name: Coxsackievirus and adenovirus receptor / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.106408 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSITTPEEMI EKAKGETAYL PCKFTLSPED QGPLDIEWLI SPADNQKVDQ VIILYSGDKI YDDYYPDLKG RVHFTSNDLK SGDASINVT NLQLSDIGTY QCKVKKAPGV ANKKIHLVVL VKPSGARCYV DGSEEIGSDF KIKCEPKEGS LPLQYEWQKL S DSQKMPTS ...String: MSITTPEEMI EKAKGETAYL PCKFTLSPED QGPLDIEWLI SPADNQKVDQ VIILYSGDKI YDDYYPDLKG RVHFTSNDLK SGDASINVT NLQLSDIGTY QCKVKKAPGV ANKKIHLVVL VKPSGARCYV DGSEEIGSDF KIKCEPKEGS LPLQYEWQKL S DSQKMPTS WLAEMTSSVI SVKNASSEYS GTYSCTVRNR VGSDQCLLRL NVVPPSNKAL EHHHHHH UniProtKB: Coxsackievirus and adenovirus receptor |
-Macromolecule #6: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 6 / Number of copies: 1 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Support film - Material: CARBON / Support film - topology: CONTINUOUS |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average exposure time: 1.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 5.0 µm / Calibrated defocus min: 1.8 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT | ||||||||||
| Output model |  PDB-7vyk: |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)