+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0943 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Enterovirus 71 in complex with NLD-22 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Inhibitor / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |   Human enterovirus 71 Human enterovirus 71 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.264 Å | |||||||||
 Authors Authors | Zhang M / Sun Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: J Med Chem / Year: 2020 Journal: J Med Chem / Year: 2020Title: Design, Synthesis, and Evaluation of Novel Enterovirus 71 Inhibitors as Therapeutic Drug Leads for the Treatment of Human Hand, Foot, and Mouth Disease. Authors: Min Zhang / Ying Wang / Wanli He / Yao Sun / Yan Guo / Weilong Zhong / Qiang Gao / Mingyang Liao / Xiangxi Wang / Yan Cai / Yu Guo / Zihe Rao /  Abstract: Human hand, foot, and mouth disease (HFMD) is a serious public health threat with high infection rates in children and infants who reside in Asia and the Pacific regions, and no effective drugs are ...Human hand, foot, and mouth disease (HFMD) is a serious public health threat with high infection rates in children and infants who reside in Asia and the Pacific regions, and no effective drugs are currently available. Enterovirus 71 (EV71) and coxsackievirus A16 are the major etiological pathogens. Based on an essential hydrophobic pocket on the viral capsid protein VP1, we designed and synthesized a series of small molecular weight compounds as inhibitors of EV71. A potential drug candidate named exhibited excellent antiviral activity (with an EC of 5.056 nM and a 100% protection rate for mice at a dose of 20 mg/kg) and low toxicity. had a favorable pharmacokinetic profile. High-resolution cryo-electron microscopy structural analysis confirmed bound to the hydrophobic pocket in VP1 to block viral infection. In general, was indicated to be a promising potential drug candidate for the treatment of HFMD. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0943.map.gz emd_0943.map.gz | 34.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0943-v30.xml emd-0943-v30.xml emd-0943.xml emd-0943.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0943.png emd_0943.png | 225.5 KB | ||
| Filedesc metadata |  emd-0943.cif.gz emd-0943.cif.gz | 5.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0943 http://ftp.pdbj.org/pub/emdb/structures/EMD-0943 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0943 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0943 | HTTPS FTP |
-Related structure data
| Related structure data |  6lqdMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0943.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0943.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human enterovirus 71
| Entire | Name:   Human enterovirus 71 Human enterovirus 71 |
|---|---|
| Components |
|
-Supramolecule #1: Human enterovirus 71
| Supramolecule | Name: Human enterovirus 71 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 / NCBI-ID: 39054 / Sci species name: Human enterovirus 71 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:   Human enterovirus 71 Human enterovirus 71 |
| Molecular weight | Theoretical: 32.727891 KDa |
| Sequence | String: GDRVADVIES SIGDSVSRAL THALPAPTGQ NTQVSSHRLD TGKVPALQAA EIGASSNASD ESMIETRCVL NSHSTAETTL DSFFSRAGL VGEIDLPLKG TTNPNGYANW DIDITGYAQM RRKVELFTYM RFDAEFTFVA CTPTGEVVPQ LLQYMFVPPG A PKPDSRES ...String: GDRVADVIES SIGDSVSRAL THALPAPTGQ NTQVSSHRLD TGKVPALQAA EIGASSNASD ESMIETRCVL NSHSTAETTL DSFFSRAGL VGEIDLPLKG TTNPNGYANW DIDITGYAQM RRKVELFTYM RFDAEFTFVA CTPTGEVVPQ LLQYMFVPPG A PKPDSRES LAWQTATNPS VFVKLSDPPA QVSVPFMSPA SAYQWFYDGY PTFGEHKQEK DLEYGAMPNN MMGTFSVRTV GT SKSKYPL VVRIYMRMKH VRAWIPRPMR NQNYLFKANP NYAGNSIKPT GASRTAITTL UniProtKB: Genome polyprotein |
-Macromolecule #2: Capsid protein VP2
| Macromolecule | Name: Capsid protein VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:   Human enterovirus 71 Human enterovirus 71 |
| Molecular weight | Theoretical: 27.74016 KDa |
| Sequence | String: SPSAEACGYS DRVAQLTIGN STITTQEAAN IIVGYGEWPS YCSDSDATAV DKPTRPDVSV NRFYTLDTKL WEKSSKGWYW KFPDVLTET GVFGQNAQFH YLYRSGFCIH VQCNASKFHQ GALLVAVLPE YVIGTVAGGT GTEDTHPPYK QTQPGADGFE L QHPYVLDA ...String: SPSAEACGYS DRVAQLTIGN STITTQEAAN IIVGYGEWPS YCSDSDATAV DKPTRPDVSV NRFYTLDTKL WEKSSKGWYW KFPDVLTET GVFGQNAQFH YLYRSGFCIH VQCNASKFHQ GALLVAVLPE YVIGTVAGGT GTEDTHPPYK QTQPGADGFE L QHPYVLDA GIPISQLTVC PHQWINLRTN NCATIIVPYI NALPFDSALN HCNFGLLVVP ISPLDYDQGA TPVIPITITL AP MCSEFAG LRQAVTQ UniProtKB: Genome polyprotein |
-Macromolecule #3: Capsid protein VP3
| Macromolecule | Name: Capsid protein VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:   Human enterovirus 71 Human enterovirus 71 |
| Molecular weight | Theoretical: 26.468225 KDa |
| Sequence | String: GFPTELKPGT NQFLTTDDGV SAPILPNFHP TPCIHIPGEV RNLLELCQVE TILEVNNVPT NATSLMERLR FPVSAQAGKG ELCAVFRAD PGRNGPWQST LLGQLCGYYT QWSGSLEVTF MFTGSFMATG KMLIAYTPPG GPLPKDRATA MLGTHVIWDF G LQSSVTLV ...String: GFPTELKPGT NQFLTTDDGV SAPILPNFHP TPCIHIPGEV RNLLELCQVE TILEVNNVPT NATSLMERLR FPVSAQAGKG ELCAVFRAD PGRNGPWQST LLGQLCGYYT QWSGSLEVTF MFTGSFMATG KMLIAYTPPG GPLPKDRATA MLGTHVIWDF G LQSSVTLV IPWISNTHYR AHARDGVFDY YTTGLVSIWY QTNYVVPIGA PNTAYIIALA AAQKNFTMKL CKDASDILQT GT IQ UniProtKB: Genome polyprotein |
-Macromolecule #4: Capsid protein VP4
| Macromolecule | Name: Capsid protein VP4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:   Human enterovirus 71 Human enterovirus 71 |
| Molecular weight | Theoretical: 7.501162 KDa |
| Sequence | String: MGSQVSTQRS GSHENSNSAT EGSTINYTTI NYYKDSYAAT AGKQSLKQDP DKFANPVKDI FTEMAAPLK UniProtKB: Genome polyprotein |
-Macromolecule #5: 1-(2-azanylpyridin-4-yl)-3-[5-[4-(5-methyl-1,2,4-oxadiazol-3-yl)p...
| Macromolecule | Name: 1-(2-azanylpyridin-4-yl)-3-[5-[4-(5-methyl-1,2,4-oxadiazol-3-yl)phenoxy]pentyl]imidazolidin-2-one type: ligand / ID: 5 / Number of copies: 1 / Formula: EQ9 |
|---|---|
| Molecular weight | Theoretical: 422.48 Da |
| Chemical component information | 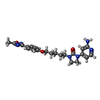 ChemComp-EQ9: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.264 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 13185 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















