[English] 日本語
 Yorodumi
Yorodumi- EMDB-9055: CryoEM structure of human enterovirus D68 expanded 1 particle (pH... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9055 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of human enterovirus D68 expanded 1 particle (pH 6.5, 4 degrees Celsius, 3 min) | |||||||||
 Map data Map data | sharpened map in which the inner density is masked out | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell membrane / cysteine-type peptidase activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / helicase activity / host cell cytoplasmic vesicle membrane / ribonucleoside triphosphate phosphatase activity ...host cell membrane / cysteine-type peptidase activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / helicase activity / host cell cytoplasmic vesicle membrane / ribonucleoside triphosphate phosphatase activity / viral capsid / nucleoside-triphosphate phosphatase / host cell / channel activity / monoatomic ion transmembrane transport / host cell cytoplasm / RNA helicase activity / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / symbiont-mediated suppression of host gene expression / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / symbiont entry into host cell / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding Similarity search - Function | |||||||||
| Biological species |  Enterovirus D68 Enterovirus D68 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.46 Å | |||||||||
 Authors Authors | Liu Y / Rossmann MG | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2018 Journal: Proc Natl Acad Sci U S A / Year: 2018Title: Molecular basis for the acid-initiated uncoating of human enterovirus D68. Authors: Yue Liu / Ju Sheng / Arno L W van Vliet / Geeta Buda / Frank J M van Kuppeveld / Michael G Rossmann /   Abstract: Enterovirus D68 (EV-D68) belongs to a group of enteroviruses that contain a single positive-sense RNA genome surrounded by an icosahedral capsid. Like common cold viruses, EV-D68 mainly causes ...Enterovirus D68 (EV-D68) belongs to a group of enteroviruses that contain a single positive-sense RNA genome surrounded by an icosahedral capsid. Like common cold viruses, EV-D68 mainly causes respiratory infections and is acid-labile. The molecular mechanism by which the acid-sensitive EV-D68 virions uncoat and deliver their genome into a host cell is unknown. Using cryoelectron microscopy (cryo-EM), we have determined the structures of the full native virion and an uncoating intermediate [the A (altered) particle] of EV-D68 at 2.2- and 2.7-Å resolution, respectively. These structures showed that acid treatment of EV-D68 leads to particle expansion, externalization of the viral protein VP1 N termini from the capsid interior, and formation of pores around the icosahedral twofold axes through which the viral RNA can exit. Moreover, because of the low stability of EV-D68, cryo-EM analyses of a mixed population of particles at neutral pH and following acid treatment demonstrated the involvement of multiple structural intermediates during virus uncoating. Among these, a previously undescribed state, the expanded 1 ("E1") particle, shows a majority of internal regions (e.g., the VP1 N termini) to be ordered as in the full native virion. Thus, the E1 particle acts as an intermediate in the transition from full native virions to A particles. Together, the present work delineates the pathway of EV-D68 uncoating and provides the molecular basis for the acid lability of EV-D68 and of the related common cold viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9055.map.gz emd_9055.map.gz | 65.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9055-v30.xml emd-9055-v30.xml emd-9055.xml emd-9055.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9055_fsc.xml emd_9055_fsc.xml | 10.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_9055.png emd_9055.png | 210.4 KB | ||
| Others |  emd_9055_half_map_1.map.gz emd_9055_half_map_1.map.gz emd_9055_half_map_2.map.gz emd_9055_half_map_2.map.gz | 21 MB 21.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9055 http://ftp.pdbj.org/pub/emdb/structures/EMD-9055 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9055 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9055 | HTTPS FTP |
-Related structure data
| Related structure data |  6mziMC  7567C  7569C  7571C  7572C  7583C  7589C  7592C  7593C  7598C  7599C  7600C  9053C  9054C  9056C  9057C  9058C  9059C  9060C  6crpC  6crrC  6crsC  6cruC  6cs3C  6cs4C  6cs5C  6cs6C  6csaC  6csgC  6cshC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9055.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9055.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map in which the inner density is masked out | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.73 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
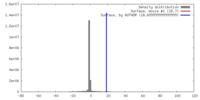
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: unsharpened half map in which the inner density is retained
| File | emd_9055_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened half map in which the inner density is retained | ||||||||||||
| Projections & Slices |
| ||||||||||||
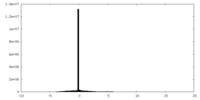
| Density Histograms |
-Half map: unsharpened half map in which the inner density is retained
| File | emd_9055_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened half map in which the inner density is retained | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Enterovirus D68
| Entire | Name:  Enterovirus D68 Enterovirus D68 |
|---|---|
| Components |
|
-Supramolecule #1: Enterovirus D68
| Supramolecule | Name: Enterovirus D68 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Viruses were grown in RD cells and treated with acidic buffer. NCBI-ID: 42789 / Sci species name: Enterovirus D68 / Sci species strain: US/MO/14-18947 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Viral protein 1
| Macromolecule | Name: Viral protein 1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterovirus D68 / Strain: US/MO/14-18947 Enterovirus D68 / Strain: US/MO/14-18947 |
| Sequence | String: IESIIKTATD TVKSEINAEL GVVPSLNAVE TGATSN TEP EEAIQTRTVI NQHGVSETLV ENFLSRAALV SKRSFEYKDH TSSTARADKN FFKWTIN TR SFVQLRRKLE LFTYLRFDAE ITILTTVAVN GSGNNTYVGL PDLTLQAMFV PTGALTPE K QDSFHWQSGS ...String: IESIIKTATD TVKSEINAEL GVVPSLNAVE TGATSN TEP EEAIQTRTVI NQHGVSETLV ENFLSRAALV SKRSFEYKDH TSSTARADKN FFKWTIN TR SFVQLRRKLE LFTYLRFDAE ITILTTVAVN GSGNNTYVGL PDLTLQAMFV PTGALTPE K QDSFHWQSGS NASVFFKISD PPARITIPFM CINSAYSVFY DGFAGFEKNG LYGINPADT IGNLCVRIVN EHQPVGFTVT VRVYMKPKHI KAWAPRPPRT LPYMSIANAN YKGKERAPNA LSAIIGNRD SVKTMPHNIV NT |
-Macromolecule #2: Viral protein 2
| Macromolecule | Name: Viral protein 2 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterovirus D68 / Strain: US/MO/14-18947 Enterovirus D68 / Strain: US/MO/14-18947 |
| Sequence | String: SPSAEACGYS DRVLQLKLGN SAIVTQEAAN YCCAYGEWPN YLPDHEAVAI D KPTQPETA TDRFYTLKSV KWETGSTGWW WKLPDALNNI GMFGQNVQHH YLYRSGFLIH VQ CNATKFH QGALLVVAIP EHQRGAHNTN TSPGFDDIMK GEEGGTFNHP YVLDDGTSLA CAT ...String: SPSAEACGYS DRVLQLKLGN SAIVTQEAAN YCCAYGEWPN YLPDHEAVAI D KPTQPETA TDRFYTLKSV KWETGSTGWW WKLPDALNNI GMFGQNVQHH YLYRSGFLIH VQ CNATKFH QGALLVVAIP EHQRGAHNTN TSPGFDDIMK GEEGGTFNHP YVLDDGTSLA CAT IFPHQW INLRTNNSAT IVLPWMNAAP MDFPLRHNQW TLAIIPVVPL GTRTTSSMVP ITVS IAPMC CEFNGLRHAI TQ |
-Macromolecule #3: Viral protein 3
| Macromolecule | Name: Viral protein 3 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterovirus D68 / Strain: US/MO/14-18947 Enterovirus D68 / Strain: US/MO/14-18947 |
| Sequence | String: GVPTYLLPGS GQFLTTDDHS SAPALPCFNP TPEMHIPGQV RNM LEVVQV ESMMEINNTE SAVGMERLKV DISALTDVDQ LLFNIPLDIQ LDGPLRNTLV GNIS RYYTH WSGSLEMTFM FCGSFMAAGK LILCYTPPGG SCPTTRETAM LGTHIVWDFG LQSSV TLII ...String: GVPTYLLPGS GQFLTTDDHS SAPALPCFNP TPEMHIPGQV RNM LEVVQV ESMMEINNTE SAVGMERLKV DISALTDVDQ LLFNIPLDIQ LDGPLRNTLV GNIS RYYTH WSGSLEMTFM FCGSFMAAGK LILCYTPPGG SCPTTRETAM LGTHIVWDFG LQSSV TLII PWISGSHYRM FNNDAKSTNA NVGYVTCFMQ TNLIVPSESS DTCSLIGFIA AKDDFS LRL MRDSPDIGQL DHLHAAEAAY Q |
-Macromolecule #4: Viral protein 4
| Macromolecule | Name: Viral protein 4 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterovirus D68 / Strain: US/MO/14-18947 Enterovirus D68 / Strain: US/MO/14-18947 |
| Sequence | String: GAQVTRQQTG THENANIATN GSHITYNQIN FYKDSYAASA SKQDFSQDPS KFTEPVVEG LKAGAPVLK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 / Details: phosphate citrate buffer |
|---|---|
| Grid | Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.02 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 298 K / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Sampling interval: 5.0 µm / Number grids imaged: 1 / Number real images: 357 / Average exposure time: 10.5 sec. / Average electron dose: 28.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 8.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-6mzi: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)