+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30848 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a human NHE1-CHP1 complex under pH 7.5 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcation-transporting ATPase complex / Sodium/Proton exchangers / regulation of the force of heart contraction by cardiac conduction / Hyaluronan degradation / regulation of cardiac muscle cell membrane potential / cellular response to electrical stimulus / potassium:proton antiporter activity / positive regulation of action potential / sodium:proton antiporter activity / maintenance of cell polarity ...cation-transporting ATPase complex / Sodium/Proton exchangers / regulation of the force of heart contraction by cardiac conduction / Hyaluronan degradation / regulation of cardiac muscle cell membrane potential / cellular response to electrical stimulus / potassium:proton antiporter activity / positive regulation of action potential / sodium:proton antiporter activity / maintenance of cell polarity / regulation of pH / sodium ion export across plasma membrane / positive regulation of calcineurin-NFAT signaling cascade / cardiac muscle cell differentiation / protein phosphatase 2B binding / response to acidic pH / intracellular sodium ion homeostasis / regulation of stress fiber assembly / cellular response to acidic pH / sodium ion import across plasma membrane / cardiac muscle cell contraction / positive regulation of mitochondrial membrane permeability / regulation of cardiac muscle contraction by calcium ion signaling / regulation of focal adhesion assembly / cellular response to antibiotic / positive regulation of cardiac muscle hypertrophy / cellular response to cold / positive regulation of the force of heart contraction / protein complex oligomerization / intercalated disc / monoatomic ion transport / response to muscle stretch / phosphatidylinositol-4,5-bisphosphate binding / cellular response to epinephrine stimulus / potassium ion transmembrane transport / T-tubule / proton transmembrane transport / regulation of intracellular pH / stem cell differentiation / cellular response to mechanical stimulus / phospholipid binding / cellular response to insulin stimulus / calcium-dependent protein binding / cell migration / lamellipodium / positive regulation of cell growth / protein-macromolecule adaptor activity / basolateral plasma membrane / cellular response to hypoxia / molecular adaptor activity / calmodulin binding / apical plasma membrane / positive regulation of apoptotic process / membrane raft / focal adhesion / negative regulation of apoptotic process / perinuclear region of cytoplasm / cell surface / positive regulation of transcription by RNA polymerase II / mitochondrion / extracellular exosome / nucleoplasm / identical protein binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
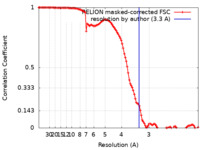
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Dong Y / Gao Y / Li B / Zhang XC / Zhao Y | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structure and mechanism of the human NHE1-CHP1 complex. Authors: Yanli Dong / Yiwei Gao / Alina Ilie / DuSik Kim / Annie Boucher / Bin Li / Xuejun C Zhang / John Orlowski / Yan Zhao /   Abstract: Sodium/proton exchanger 1 (NHE1) is an electroneutral secondary active transporter present on the plasma membrane of most mammalian cells and plays critical roles in regulating intracellular pH and ...Sodium/proton exchanger 1 (NHE1) is an electroneutral secondary active transporter present on the plasma membrane of most mammalian cells and plays critical roles in regulating intracellular pH and volume homeostasis. Calcineurin B-homologous protein 1 (CHP1) is an obligate binding partner that promotes NHE1 biosynthetic maturation, cell surface expression and pH-sensitivity. Dysfunctions of either protein are associated with neurological disorders. Here, we elucidate structures of the human NHE1-CHP1 complex in both inward- and inhibitor (cariporide)-bound outward-facing conformations. We find that NHE1 assembles as a symmetrical homodimer, with each subunit undergoing an elevator-like conformational change during cation exchange. The cryo-EM map reveals the binding site for the NHE1 inhibitor cariporide, illustrating how inhibitors block transport activity. The CHP1 molecule differentially associates with these two conformational states of each NHE1 monomer, and this association difference probably underlies the regulation of NHE1 pH-sensitivity by CHP1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30848.map.gz emd_30848.map.gz | 4.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30848-v30.xml emd-30848-v30.xml emd-30848.xml emd-30848.xml | 13.1 KB 13.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30848_fsc.xml emd_30848_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_30848.png emd_30848.png | 198.3 KB | ||
| Filedesc metadata |  emd-30848.cif.gz emd-30848.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30848 http://ftp.pdbj.org/pub/emdb/structures/EMD-30848 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30848 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30848 | HTTPS FTP |
-Validation report
| Summary document |  emd_30848_validation.pdf.gz emd_30848_validation.pdf.gz | 378.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30848_full_validation.pdf.gz emd_30848_full_validation.pdf.gz | 377.8 KB | Display | |
| Data in XML |  emd_30848_validation.xml.gz emd_30848_validation.xml.gz | 11.6 KB | Display | |
| Data in CIF |  emd_30848_validation.cif.gz emd_30848_validation.cif.gz | 15.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30848 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30848 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30848 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30848 | HTTPS FTP |
-Related structure data
| Related structure data |  7dswMC  7dsvC  7dsxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_30848.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30848.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human NHE1-CHP1 complex under pH 7.5
| Entire | Name: Human NHE1-CHP1 complex under pH 7.5 |
|---|---|
| Components |
|
-Supramolecule #1: Human NHE1-CHP1 complex under pH 7.5
| Supramolecule | Name: Human NHE1-CHP1 complex under pH 7.5 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Calcineurin B homologous protein 1 (CHP1) was not successfully resolved in this map because of conformational heterogeneity. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 230 KDa |
-Macromolecule #1: Sodium/hydrogen exchanger 1
| Macromolecule | Name: Sodium/hydrogen exchanger 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 46.610219 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: KAFPVLGIDY THVRTPFEIS LWILLACLMK IGFHVIPTIS SIVPESCLLI VVGLLVGGLI KGVGETPPFL QSDVFFLFLL PPIILDAGY FLPLRQFTEN LGTILIFAVV GTLWNAFFLG GLMYAVCLVG GEQINNIGLL DNLLFGSIIS AVDPVAVLAV F EEIHINEL ...String: KAFPVLGIDY THVRTPFEIS LWILLACLMK IGFHVIPTIS SIVPESCLLI VVGLLVGGLI KGVGETPPFL QSDVFFLFLL PPIILDAGY FLPLRQFTEN LGTILIFAVV GTLWNAFFLG GLMYAVCLVG GEQINNIGLL DNLLFGSIIS AVDPVAVLAV F EEIHINEL LHILVFGESL LNDAVTVVLY HLFEEFANYE HVGIVDIFLG FLSFFVVALG GVLVGVVYGV IAAFTSRFTS HI RVIEPLF VFLYSYMAYL SAELFHLSGI MALIASGVVM RPYVEANISH KSHTTIKYFL KMWSSVSETL IFIFLGVSTV AGS HHWNWT FVISTLLFCL IARVLGVLGL TWFINKFRIV KLTPKDQFII AYGGLRGAIA FSLGYLLDKK HFPMCDLFLT AIIT VIFFT VFVQGMTIRP LVDLL UniProtKB: Sodium/hydrogen exchanger 1 |
-Macromolecule #2: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
| Macromolecule | Name: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine / type: ligand / ID: 2 / Number of copies: 12 / Formula: LBN |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-LBN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL |
|---|---|
| Buffer | pH: 6.5 |
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
| Details | The NHE1-CHP1 complex was reconstituted into lipid nanodiscs. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-32 / Number grids imaged: 1 / Number real images: 2462 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 13000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7dsw: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)