+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30023 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tetrameric structure of AtCRY2 PHR domain | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
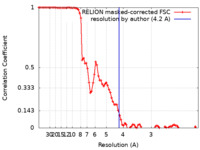
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Shao K / Zhang X / Zhang P | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2020 Journal: Nat Struct Mol Biol / Year: 2020Title: The oligomeric structures of plant cryptochromes. Authors: Kai Shao / Xue Zhang / Xu Li / Yahui Hao / Xiaowei Huang / Miaolian Ma / Minhua Zhang / Fang Yu / Hongtao Liu / Peng Zhang /  Abstract: Cryptochromes (CRYs) are a group of evolutionarily conserved flavoproteins found in many organisms. In plants, the well-studied CRY photoreceptor, activated by blue light, plays essential roles in ...Cryptochromes (CRYs) are a group of evolutionarily conserved flavoproteins found in many organisms. In plants, the well-studied CRY photoreceptor, activated by blue light, plays essential roles in plant growth and development. However, the mechanism of activation remains largely unknown. Here, we determined the oligomeric structures of the blue-light-perceiving PHR domain of Zea mays CRY1 and an Arabidopsis CRY2 constitutively active mutant. The structures form dimers and tetramers whose functional importance is examined in vitro and in vivo with Arabidopsis CRY2. Structure-based analysis suggests that blue light may be perceived by CRY to cause conformational changes, whose precise nature remains to be determined, leading to oligomerization that is essential for downstream signaling. This photoactivation mechanism may be widely used by plant CRYs. Our study reveals a molecular mechanism of plant CRY activation and also paves the way for design of CRY as a more efficient optical switch. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30023.map.gz emd_30023.map.gz | 3.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30023-v30.xml emd-30023-v30.xml emd-30023.xml emd-30023.xml | 7.4 KB 7.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30023_fsc.xml emd_30023_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_30023.png emd_30023.png | 46.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30023 http://ftp.pdbj.org/pub/emdb/structures/EMD-30023 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30023 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30023 | HTTPS FTP |
-Validation report
| Summary document |  emd_30023_validation.pdf.gz emd_30023_validation.pdf.gz | 336.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30023_full_validation.pdf.gz emd_30023_full_validation.pdf.gz | 335.7 KB | Display | |
| Data in XML |  emd_30023_validation.xml.gz emd_30023_validation.xml.gz | 10.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30023 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30023 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30023 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30023 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_30023.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30023.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.061 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : tetrameric structure of AtCRY2 PHR domain
| Entire | Name: tetrameric structure of AtCRY2 PHR domain |
|---|---|
| Components |
|
-Supramolecule #1: tetrameric structure of AtCRY2 PHR domain
| Supramolecule | Name: tetrameric structure of AtCRY2 PHR domain / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 49.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)