+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human liver glucosidase II | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | glucosidase II / GANAB / glycosyl hydrolase 31 family / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationglucan 1,3-alpha-glucosidase activity / Glc2Man9GlcNAc2 oligosaccharide glucosidase activity / mannosyl-oligosaccharide alpha-1,3-glucosidase / Calnexin/calreticulin cycle / alpha-glucosidase activity / glucosidase II complex / N-glycan processing / Maturation of spike protein / Regulation of CDH1 posttranslational processing and trafficking to plasma membrane / Advanced glycosylation endproduct receptor signaling ...glucan 1,3-alpha-glucosidase activity / Glc2Man9GlcNAc2 oligosaccharide glucosidase activity / mannosyl-oligosaccharide alpha-1,3-glucosidase / Calnexin/calreticulin cycle / alpha-glucosidase activity / glucosidase II complex / N-glycan processing / Maturation of spike protein / Regulation of CDH1 posttranslational processing and trafficking to plasma membrane / Advanced glycosylation endproduct receptor signaling / protein kinase C binding / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / Post-translational protein phosphorylation / phosphoprotein binding / liver development / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / melanosome / carbohydrate binding / Maturation of spike protein / carbohydrate metabolic process / transmembrane transporter binding / intracellular signal transduction / endoplasmic reticulum lumen / intracellular membrane-bounded organelle / calcium ion binding / endoplasmic reticulum / Golgi apparatus / RNA binding / extracellular exosome / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
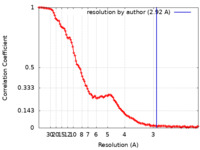
| Method | single particle reconstruction / cryo EM / Resolution: 2.92 Å | |||||||||
 Authors Authors | Su C / Lyu M / Zhang Z / Yu EW | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2023 Journal: Cell Rep / Year: 2023Title: High-resolution structural-omics of human liver enzymes. Authors: Chih-Chia Su / Meinan Lyu / Zhemin Zhang / Masaru Miyagi / Wei Huang / Derek J Taylor / Edward W Yu /  Abstract: We applied raw human liver microsome lysate to a holey carbon grid and used cryo-electron microscopy (cryo-EM) to define its composition. From this sample we identified and simultaneously determined ...We applied raw human liver microsome lysate to a holey carbon grid and used cryo-electron microscopy (cryo-EM) to define its composition. From this sample we identified and simultaneously determined high-resolution structural information for ten unique human liver enzymes involved in diverse cellular processes. Notably, we determined the structure of the endoplasmic bifunctional protein H6PD, where the N- and C-terminal domains independently possess glucose-6-phosphate dehydrogenase and 6-phosphogluconolactonase enzymatic activity, respectively. We also obtained the structure of heterodimeric human GANAB, an ER glycoprotein quality-control machinery that contains a catalytic α subunit and a noncatalytic β subunit. In addition, we observed a decameric peroxidase, PRDX4, which directly contacts a disulfide isomerase-related protein, ERp46. Structural data suggest that several glycosylations, bound endogenous compounds, and ions associate with these human liver enzymes. These results highlight the importance of cryo-EM in facilitating the elucidation of human organ proteomics at the atomic level. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28262.map.gz emd_28262.map.gz | 97.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28262-v30.xml emd-28262-v30.xml emd-28262.xml emd-28262.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28262_fsc.xml emd_28262_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_28262.png emd_28262.png | 144.5 KB | ||
| Masks |  emd_28262_msk_1.map emd_28262_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-28262.cif.gz emd-28262.cif.gz | 6.4 KB | ||
| Others |  emd_28262_additional_1.map.gz emd_28262_additional_1.map.gz emd_28262_half_map_1.map.gz emd_28262_half_map_1.map.gz emd_28262_half_map_2.map.gz emd_28262_half_map_2.map.gz | 3 MB 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28262 http://ftp.pdbj.org/pub/emdb/structures/EMD-28262 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28262 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28262 | HTTPS FTP |
-Related structure data
| Related structure data |  8emrMC  7uzmC  8ekwC  8ekyC  8em2C  8emsC  8emtC  8eneC  8eojC  8eorC  23434 M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28262.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28262.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
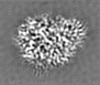
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
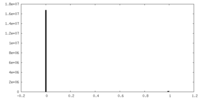
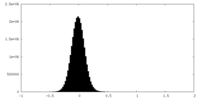
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28262_msk_1.map emd_28262_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
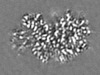
| Projections & Slices |
| ||||||||||||
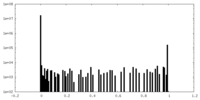
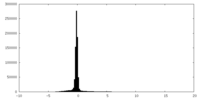
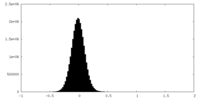
| Density Histograms |
-Additional map: density modification map
| File | emd_28262_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | density modification map | ||||||||||||
| Projections & Slices |
| ||||||||||||
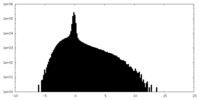
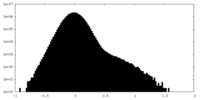
| Density Histograms |
-Half map: #2
| File | emd_28262_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_28262_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : glucosidase II
| Entire | Name: glucosidase II |
|---|---|
| Components |
|
-Supramolecule #1: glucosidase II
| Supramolecule | Name: glucosidase II / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Neutral alpha-glucosidase AB
| Macromolecule | Name: Neutral alpha-glucosidase AB / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: mannosyl-oligosaccharide alpha-1,3-glucosidase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: liver Homo sapiens (human) / Organ: liver |
| Molecular weight | Theoretical: 106.997828 KDa |
| Sequence | String: MAAVAAVAAR RRRSWASLVL AFLGVCLGIT LAVDRSNFKT CEESSFCKRQ RSIRPGLSPY RALLDSLQLG PDSLTVHLIH EVTKVLLVL ELQGLQKNMT RFRIDELEPR RPRYRVPDVL VADPPIARLS VSGRDENSVE LTMAEGPYKI ILTARPFRLD L LEDRSLLL ...String: MAAVAAVAAR RRRSWASLVL AFLGVCLGIT LAVDRSNFKT CEESSFCKRQ RSIRPGLSPY RALLDSLQLG PDSLTVHLIH EVTKVLLVL ELQGLQKNMT RFRIDELEPR RPRYRVPDVL VADPPIARLS VSGRDENSVE LTMAEGPYKI ILTARPFRLD L LEDRSLLL SVNARGLLEF EHQRAPRVSQ GSKDPAEGDG AQPEETPRDG DKPEETQGKA EKDEPGAWEE TFKTHSDSKP YG PMSVGLD FSLPGMEHVY GIPEHADNLR LKVTEGGEPY RLYNLDVFQY ELYNPMALYG SVPVLLAHNP HRDLGIFWLN AAE TWVDIS SNTAGKTLFG KMMDYLQGSG ETPQTDVRWM SETGIIDVFL LLGPSISDVF RQYASLTGTQ ALPPLFSLGY HQSR WNYRD EADVLEVDQG FDDHNLPCDV IWLDIEHADG KRYFTWDPSR FPQPRTMLER LASKRRKLVA IVDPHIKVDS GYRVH EELR NLGLYVKTRD GSDYEGWCWP GSAGYPDFTN PTMRAWWANM FSYDNYEGSA PNLFVWNDMN EPSVFNGPEV TMLKDA QHY GGWEHRDVHN IYGLYVHMAT ADGLRQRSGG MERPFVLARA FFAGSQRFGA VWTGDNTAEW DHLKISIPMC LSLGLVG LS FCGADVGGFF KNPEPELLVR WYQMGAYQPF FRAHAHLDTG RREPWLLPSQ HNDIIRDALG QRYSLLPFWY TLLYQAHR E GIPVMRPLWV QYPQDVTTFN IDDQYLLGDA LLVHPVSDSG AHGVQVYLPG QGEVWYDIQS YQKHHGPQTL YLPVTLSSI PVFQRGGTIV PRWMRVRRSS ECMKDDPITL FVALSPQGTA QGELFLDDGH TFNYQTRQEF LLRRFSFSGN TLVSSSADPE GHFETPIWI ERVVIIGAGK PAAVVLQTKG SPESRLSFQH DPETSVLVLR KPGINVASDW SIHLR UniProtKB: Neutral alpha-glucosidase AB |
-Macromolecule #2: Glucosidase 2 subunit beta
| Macromolecule | Name: Glucosidase 2 subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: liver Homo sapiens (human) / Organ: liver |
| Molecular weight | Theoretical: 59.485223 KDa |
| Sequence | String: MLLPLLLLLP MCWAVEVKRP RGVSLTNHHF YDESKPFTCL DGSATIPFDQ VNDDYCDCKD GSDEPGTAAC PNGSFHCTNT GYKPLYIPS NRVNDGVCDC CDGTDEYNSG VICENTCKEK GRKERESLQQ MAEVTREGFR LKKILIEDWK KAREEKQKKL I ELQAGKKS ...String: MLLPLLLLLP MCWAVEVKRP RGVSLTNHHF YDESKPFTCL DGSATIPFDQ VNDDYCDCKD GSDEPGTAAC PNGSFHCTNT GYKPLYIPS NRVNDGVCDC CDGTDEYNSG VICENTCKEK GRKERESLQQ MAEVTREGFR LKKILIEDWK KAREEKQKKL I ELQAGKKS LEDQVEMLRT VKEEAEKPER EAKEQHQKLW EEQLAAAKAQ QEQELAADAF KELDDDMDGT VSVTELQTHP EL DTDGDGA LSEAEAQALL SGDTQTDATS FYDRVWAAIR DKYRSEALPT DLPAPSAPDL TEPKEEQPPV PSSPTEEEEE EEE EEEEEA EEEEEEEDSE EAPPPLSPPQ PASPAEEDKM PPYDEQTQAF IDAAQEARNK FEEAERSLKD MEESIRNLEQ EISF DFGPN GEFAYLYSQC YELTTNEYVY RLCPFKLVSQ KPKLGGSPTS LGTWGSWIGP DHDKFSAMKY EQGTGCWQGP NRSTT VRLL CGKETMVTST TEPSRCEYLM ELMTPAACPE PPPEAPTEDD HDEL UniProtKB: Glucosidase 2 subunit beta |
-Macromolecule #4: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 41.25 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)