[English] 日本語
 Yorodumi
Yorodumi- EMDB-2621: PRE-translocational (classical-1) 80S ribosomal state of Regulati... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2621 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | PRE-translocational (classical-1) 80S ribosomal state of Regulation of the mammalian elongation cycle by 40S subunit rolling: a eukaryotic-specific ribosome rearrangement | |||||||||
 Map data Map data | reconstruction of pre-translocation (classical-1) state of 80S ribosome | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | translation / mammalian 80S ribosome / elongation cycle / pre-translocational state / tRNA selection / cryo-electron microscopy | |||||||||
| Biological species |    | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.6 Å | |||||||||
 Authors Authors | Budkevich TV / Giesebrecht J / Behrmann E / Loerke J / Ramrath D / Mielke T / Ismer J / Hildebrand P / Tung C-S / Nierhaus KH ...Budkevich TV / Giesebrecht J / Behrmann E / Loerke J / Ramrath D / Mielke T / Ismer J / Hildebrand P / Tung C-S / Nierhaus KH / Sanbonmatsu KY / Spahn CMT | |||||||||
 Citation Citation |  Journal: Cell / Year: 2014 Journal: Cell / Year: 2014Title: Regulation of the mammalian elongation cycle by subunit rolling: a eukaryotic-specific ribosome rearrangement. Authors: Tatyana V Budkevich / Jan Giesebrecht / Elmar Behrmann / Justus Loerke / David J F Ramrath / Thorsten Mielke / Jochen Ismer / Peter W Hildebrand / Chang-Shung Tung / Knud H Nierhaus / ...Authors: Tatyana V Budkevich / Jan Giesebrecht / Elmar Behrmann / Justus Loerke / David J F Ramrath / Thorsten Mielke / Jochen Ismer / Peter W Hildebrand / Chang-Shung Tung / Knud H Nierhaus / Karissa Y Sanbonmatsu / Christian M T Spahn /    Abstract: The extent to which bacterial ribosomes and the significantly larger eukaryotic ribosomes share the same mechanisms of ribosomal elongation is unknown. Here, we present subnanometer resolution ...The extent to which bacterial ribosomes and the significantly larger eukaryotic ribosomes share the same mechanisms of ribosomal elongation is unknown. Here, we present subnanometer resolution cryoelectron microscopy maps of the mammalian 80S ribosome in the posttranslocational state and in complex with the eukaryotic eEF1A⋅Val-tRNA⋅GMPPNP ternary complex, revealing significant differences in the elongation mechanism between bacteria and mammals. Surprisingly, and in contrast to bacterial ribosomes, a rotation of the small subunit around its long axis and orthogonal to the well-known intersubunit rotation distinguishes the posttranslocational state from the classical pretranslocational state ribosome. We term this motion "subunit rolling." Correspondingly, a mammalian decoding complex visualized in substates before and after codon recognition reveals structural distinctions from the bacterial system. These findings suggest how codon recognition leads to GTPase activation in the mammalian system and demonstrate that in mammalia subunit rolling occurs during tRNA selection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2621.map.gz emd_2621.map.gz | 152.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2621-v30.xml emd-2621-v30.xml emd-2621.xml emd-2621.xml | 12.6 KB 12.6 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2621_500x500.tif EMD-2621_500x500.tif | 732.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2621 http://ftp.pdbj.org/pub/emdb/structures/EMD-2621 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2621 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2621 | HTTPS FTP |
-Validation report
| Summary document |  emd_2621_validation.pdf.gz emd_2621_validation.pdf.gz | 234 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2621_full_validation.pdf.gz emd_2621_full_validation.pdf.gz | 233.2 KB | Display | |
| Data in XML |  emd_2621_validation.xml.gz emd_2621_validation.xml.gz | 7.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2621 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2621 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2621 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2621 | HTTPS FTP |
-Related structure data
| Related structure data |  2620C  2622C  2623C  2624C  4cxgC  4cxhC  4ujeC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2621.map.gz / Format: CCP4 / Size: 173.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2621.map.gz / Format: CCP4 / Size: 173.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | reconstruction of pre-translocation (classical-1) state of 80S ribosome | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.26 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
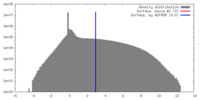
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Pre-translocational state (classical-1) of mammalian 80S ribosome...
| Entire | Name: Pre-translocational state (classical-1) of mammalian 80S ribosome with three tRNAs and mRNA bound |
|---|---|
| Components |
|
-Supramolecule #1000: Pre-translocational state (classical-1) of mammalian 80S ribosome...
| Supramolecule | Name: Pre-translocational state (classical-1) of mammalian 80S ribosome with three tRNAs and mRNA bound type: sample / ID: 1000 Details: 80S was re-associated and the sample prepared in vitro from individual components Oligomeric state: one 80S binds three tRNAs and one mRNA / Number unique components: 5 |
|---|---|
| Molecular weight | Theoretical: 4.5 MDa |
-Supramolecule #1: Re-associated 80S
| Supramolecule | Name: Re-associated 80S / type: complex / ID: 1 / Name.synonym: 80S ribosome / Recombinant expression: No / Ribosome-details: ribosome-eukaryote: ALL |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 4.5 MDa |
-Macromolecule #1: transfer RNA
| Macromolecule | Name: transfer RNA / type: rna / ID: 1 / Name.synonym: tRNA / Details: tRNAPhe / Classification: TRANSFER / Structure: DOUBLE HELIX / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25 KDa |
-Macromolecule #2: transfer RNA
| Macromolecule | Name: transfer RNA / type: rna / ID: 2 / Name.synonym: tRNA / Details: tRNALys, isoaceptor 3 / Classification: TRANSFER / Structure: DOUBLE HELIX / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25 KDa |
-Macromolecule #3: transfer RNA
| Macromolecule | Name: transfer RNA / type: rna / ID: 3 / Name.synonym: tRNA / Classification: TRANSFER / Structure: DOUBLE HELIX / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25 KDa |
-Macromolecule #4: messenger RNA
| Macromolecule | Name: messenger RNA / type: rna / ID: 4 / Name.synonym: mRNA / Classification: OTHER / Structure: SINGLE STRANDED / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.1 KDa |
| Sequence | String: GGGAAAAGAA AAGAAAAGAA AAUGUUCAAA GAAAAGAAAA GAAAU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.38 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20 mM Hepes, 5 mM MgCl2, 100 mM NH4Cl, 6 mM beta-mercaptoethanol, 0.8 mM spermidine, 0.6 mM spermine |
| Grid | Details: Quantifoil grids with additional continuous carbon support. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Instrument: FEI VITROBOT MARK II / Method: blot 2/4 sec before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 7 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 200,000 times magnification |
| Details | minimal dose system |
| Date | Jun 2, 2008 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OTHER / Number real images: 826 / Average electron dose: 20 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 65520 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 39000 |
| Sample stage | Specimen holder: Nitrogen cooled / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The particles were selected using SIGNATURE and processed by using SPIDER and SPARX. |
|---|---|
| CTF correction | Details: defocus group |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 7.6 Å / Resolution method: OTHER / Software - Name: Spider, Sparx / Number images used: 66618 |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)