+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11064 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of canine Sec61 inhibited by mycolactone | |||||||||
 Map data Map data | Experimental map used for model building | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Sec61 / mycolactone / ribosome-translocon complex / translocation inhibitor / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationInsertion of tail-anchored proteins into the endoplasmic reticulum membrane / membrane docking / pronephric nephron development / cotranslational protein targeting to membrane / Ssh1 translocon complex / Sec61 translocon complex / protein insertion into ER membrane / protein targeting to ER / SRP-dependent cotranslational protein targeting to membrane, translocation / signal sequence binding ...Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / membrane docking / pronephric nephron development / cotranslational protein targeting to membrane / Ssh1 translocon complex / Sec61 translocon complex / protein insertion into ER membrane / protein targeting to ER / SRP-dependent cotranslational protein targeting to membrane, translocation / signal sequence binding / post-translational protein targeting to membrane, translocation / protein transmembrane transporter activity / phospholipid binding / ribosome binding / endoplasmic reticulum membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
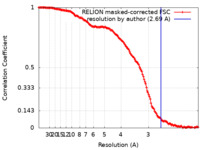
| Method | single particle reconstruction / cryo EM / Resolution: 2.69 Å | |||||||||
 Authors Authors | Gerard SF / Higgins MK | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2020 Journal: Mol Cell / Year: 2020Title: Structure of the Inhibited State of the Sec Translocon. Authors: Samuel F Gérard / Belinda S Hall / Afroditi M Zaki / Katherine A Corfield / Peter U Mayerhofer / Catia Costa / Daniel K Whelligan / Philip C Biggin / Rachel E Simmonds / Matthew K Higgins /  Abstract: Protein secretion in eukaryotes and prokaryotes involves a universally conserved protein translocation channel formed by the Sec61 complex. Unrelated small-molecule natural products and synthetic ...Protein secretion in eukaryotes and prokaryotes involves a universally conserved protein translocation channel formed by the Sec61 complex. Unrelated small-molecule natural products and synthetic compounds inhibit Sec61 with differential effects for different substrates or for Sec61 from different organisms, making this a promising target for therapeutic intervention. To understand the mode of inhibition and provide insight into the molecular mechanism of this dynamic translocon, we determined the structure of mammalian Sec61 inhibited by the Mycobacterium ulcerans exotoxin mycolactone via electron cryo-microscopy. Unexpectedly, the conformation of inhibited Sec61 is optimal for substrate engagement, with mycolactone wedging open the cytosolic side of the lateral gate. The inability of mycolactone-inhibited Sec61 to effectively transport substrate proteins implies that signal peptides and transmembrane domains pass through the site occupied by mycolactone. This provides a foundation for understanding the molecular mechanism of Sec61 inhibitors and reveals novel features of translocon function and dynamics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11064.map.gz emd_11064.map.gz | 51.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11064-v30.xml emd-11064-v30.xml emd-11064.xml emd-11064.xml | 23.5 KB 23.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11064_fsc.xml emd_11064_fsc.xml | 14.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_11064.png emd_11064.png | 531.1 KB | ||
| Masks |  emd_11064_msk_1.map emd_11064_msk_1.map | 282.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-11064.cif.gz emd-11064.cif.gz | 6.3 KB | ||
| Others |  emd_11064_additional_1.map.gz emd_11064_additional_1.map.gz emd_11064_additional_2.map.gz emd_11064_additional_2.map.gz emd_11064_half_map_1.map.gz emd_11064_half_map_1.map.gz emd_11064_half_map_2.map.gz emd_11064_half_map_2.map.gz | 36.2 MB 324.1 KB 224.8 MB 224.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11064 http://ftp.pdbj.org/pub/emdb/structures/EMD-11064 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11064 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11064 | HTTPS FTP |
-Related structure data
| Related structure data |  6z3tMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11064.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11064.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Experimental map used for model building | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.033 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11064_msk_1.map emd_11064_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
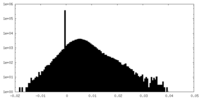
| Density Histograms |
-Additional map: Map for mycolactone-free control sample
| File | emd_11064_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map for mycolactone-free control sample | ||||||||||||
| Projections & Slices |
| ||||||||||||
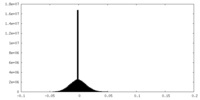
| Density Histograms |
-Additional map: Map for mycolactone-bound sample filtered to 5A resolution,...
| File | emd_11064_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map for mycolactone-bound sample filtered to 5A resolution, using for Figure 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
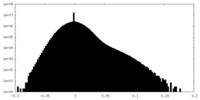
| Density Histograms |
-Half map: Half Map 1
| File | emd_11064_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
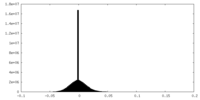
| Density Histograms |
-Half map: Half Map 2
| File | emd_11064_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ribosome-translocon complexes from canine ER microsomes, bound to...
| Entire | Name: Ribosome-translocon complexes from canine ER microsomes, bound to mycolactone |
|---|---|
| Components |
|
-Supramolecule #1: Ribosome-translocon complexes from canine ER microsomes, bound to...
| Supramolecule | Name: Ribosome-translocon complexes from canine ER microsomes, bound to mycolactone type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Protein transport protein Sec61 subunit alpha isoform 1
| Macromolecule | Name: Protein transport protein Sec61 subunit alpha isoform 1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 52.279379 KDa |
| Sequence | String: MAIKFLEVIK PFCVILPEIQ KPERKIQFKE KVLWTAITLF IFLVCCQIPL FGIMSSDSAD PFYWMRVILA SNRGTLMELG ISPIVTSGL IMQLLAGAKI IEVGDTPKDR ALFNGAQKLF GMIITIGQSI VYVMTGMYGD PSEMGAGICL LITIQLFVAG L IVLLLDEL ...String: MAIKFLEVIK PFCVILPEIQ KPERKIQFKE KVLWTAITLF IFLVCCQIPL FGIMSSDSAD PFYWMRVILA SNRGTLMELG ISPIVTSGL IMQLLAGAKI IEVGDTPKDR ALFNGAQKLF GMIITIGQSI VYVMTGMYGD PSEMGAGICL LITIQLFVAG L IVLLLDEL LQKGYGLGSG ISLFIATNIC ETIVWKAFSP TTVNTGRGME FEGAIIALFH LLATRTDKVR ALREAFYRQN LP NLMNLIA TIFVFAVVIY FQGFRVDLPI KSARYRGQYN TYPIKLFYTS NIPIILQSAL VSNLYVISQM LSARFSGNLL VSL LGTWSD TSSGGPARAY PVGGLCHYLS PPESFGSVLE DPVHAVVYIV FMLGSCAFFS KTWIEVSGSS AKDVAKQLKE QQMV MRGHR ETSMVHELNR YIPTAAAFGG LCIGALSVLA DFLGAIGSGT GILLAVTIIY QYFEIFVKEQ SEVGSMGALL F UniProtKB: Protein transport protein Sec61 subunit alpha isoform 1 |
-Macromolecule #2: Protein transport protein Sec61 subunit gamma
| Macromolecule | Name: Protein transport protein Sec61 subunit gamma / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.752325 KDa |
| Sequence | String: MDQVMQFVEP SRQFVKDSIR LVKRCTKPDR KEFQKIAMAT AIGFAIMGFI GFFVKLIHIP INNIIVGG UniProtKB: Protein transport protein Sec61 subunit gamma |
-Macromolecule #3: Protein transport protein Sec61 subunit beta
| Macromolecule | Name: Protein transport protein Sec61 subunit beta / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.720111 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK) |
-Macromolecule #4: [(6~{S},7~{S},9~{Z},12~{R})-12-[(~{Z},2~{S},6~{R},7~{R},9~{R})-4,...
| Macromolecule | Name: [(6~{S},7~{S},9~{Z},12~{R})-12-[(~{Z},2~{S},6~{R},7~{R},9~{R})-4,6-dimethyl-7,9-bis(oxidanyl)dec-4-en-2-yl]-7,9-dimethyl-2-oxidanylidene-1-oxacyclododec-9-en-6-yl] ...Name: [(6~{S},7~{S},9~{Z},12~{R})-12-[(~{Z},2~{S},6~{R},7~{R},9~{R})-4,6-dimethyl-7,9-bis(oxidanyl)dec-4-en-2-yl]-7,9-dimethyl-2-oxidanylidene-1-oxacyclododec-9-en-6-yl] (2~{E},4~{E},6~{E},8~{E},10~{E},12~{S},13~{S},15~{S})-4,6,10-trimethyl-12,13,15-tris(oxidanyl)hexadeca-2,4,6,8,10-pentaenoate type: ligand / ID: 4 / Number of copies: 1 / Formula: Q6B |
|---|---|
| Molecular weight | Theoretical: 743.021 Da |
| Chemical component information | 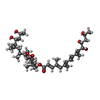 ChemComp-Q6B: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: GRAPHENE OXIDE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: Blot time 5 seconds Blot force -15 Incubation time 15 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 49.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)