+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-25905 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of W6 possum enterovirus | |||||||||
 Map data Map data | Enterovirus EV-F4 - isolated from Bushtail Possum | |||||||||
 Sample Sample | Enterovirus != Possum enterovirus W6 Enterovirus
| |||||||||
 Keywords Keywords | enterovirus / icosahedral / VIRUS | |||||||||
| Biological species |  Possum enterovirus W6 Possum enterovirus W6 | |||||||||
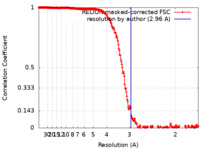
| Method | single particle reconstruction / cryo EM / Resolution: 2.96 Å | |||||||||
 Authors Authors | Wang I / Jayawardena N | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: Viruses / Year: 2022 Journal: Viruses / Year: 2022Title: Cryo-EM Structure of a Possum Enterovirus. Authors: Ivy Wang / Sandeep K Gupta / Guillaume Ems / Nadishka Jayawardena / Mike Strauss / Mihnea Bostina /     Abstract: Enteroviruses (EVs) represent a substantial concern to global health. Here, we present the cryo-EM structure of a non-human enterovirus, EV-F4, isolated from the Australian brushtail possum to assess ...Enteroviruses (EVs) represent a substantial concern to global health. Here, we present the cryo-EM structure of a non-human enterovirus, EV-F4, isolated from the Australian brushtail possum to assess the structural diversity of these picornaviruses. The capsid structure, determined to ~3 Å resolution by single particle analysis, exhibits a largely smooth surface, similar to EV-F3 (formerly BEV-2). Although the cellular receptor is not known, the absence of charged residues on the outer surface of the canyon suggest a different receptor type than for EV-F3. Density for the pocket factor is clear, with the entrance to the pocket being smaller than for other enteroviruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25905.map.gz emd_25905.map.gz | 920.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25905-v30.xml emd-25905-v30.xml emd-25905.xml emd-25905.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25905_fsc.xml emd_25905_fsc.xml | 22.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_25905.png emd_25905.png | 117.4 KB | ||
| Filedesc metadata |  emd-25905.cif.gz emd-25905.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25905 http://ftp.pdbj.org/pub/emdb/structures/EMD-25905 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25905 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25905 | HTTPS FTP |
-Related structure data
| Related structure data |  7thxMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25905.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25905.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Enterovirus EV-F4 - isolated from Bushtail Possum | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.857 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Enterovirus
| Entire | Name:  Enterovirus Enterovirus |
|---|---|
| Components |
|
-Supramolecule #1: Possum enterovirus W6
| Supramolecule | Name: Possum enterovirus W6 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 / NCBI-ID: 263532 / Sci species name: Possum enterovirus W6 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Possum enterovirus W6 Possum enterovirus W6 |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Possum enterovirus W6 Possum enterovirus W6 |
| Molecular weight | Theoretical: 30.311832 KDa |
| Sequence | String: GETGQLIKDT IKNTVENTVE STHSISTEAT PALQAAETGA TSNASDESMI ETRNVVNTHG VAETSLEGFY GRAGLVAMFT TEGGIRSWY INFGKYVQLR AKLELLTYAR FDVEFTIVAQ VVDEQAKVKD FNVDYQVMYV PPGASAPDGQ DSFQWQSSCN P SVISNTGL ...String: GETGQLIKDT IKNTVENTVE STHSISTEAT PALQAAETGA TSNASDESMI ETRNVVNTHG VAETSLEGFY GRAGLVAMFT TEGGIRSWY INFGKYVQLR AKLELLTYAR FDVEFTIVAQ VVDEQAKVKD FNVDYQVMYV PPGASAPDGQ DSFQWQSSCN P SVISNTGL PPARVSVPFM SSANAYSFSY DGYIQFGDTS SSSYGILPIH YLGQLVVRTC EDLDSARLRV RIYAKPKHMR GW IPRSPRM RPYVSRFTGI YTDAPSFCVN RESITTAG |
-Macromolecule #2: Capsid protein VP2
| Macromolecule | Name: Capsid protein VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Possum enterovirus W6 Possum enterovirus W6 |
| Molecular weight | Theoretical: 26.577887 KDa |
| Sequence | String: SAEACGYSDR VAQLTLGNST ITTQEAANIV VAYGRWPSGL RDTDATAVDK PTQPGVSAER FYTLPSVQWT TSFTGHYWKL PDALSDLGL FGQNLQFHYL YRGGWAIHVQ CNATKFHQGT LLVVAVPEHK IQAQSNPSFG RTNPGEAGAA CQFPFTFEDG T ALGNALIY ...String: SAEACGYSDR VAQLTLGNST ITTQEAANIV VAYGRWPSGL RDTDATAVDK PTQPGVSAER FYTLPSVQWT TSFTGHYWKL PDALSDLGL FGQNLQFHYL YRGGWAIHVQ CNATKFHQGT LLVVAVPEHK IQAQSNPSFG RTNPGEAGAA CQFPFTFEDG T ALGNALIY PHQWINLRTN NSATLVLPYV NAIPMDSGIK HNNWTLLVIP IVPLEYAVGA TTFVPITVTI APMCTEYNGL RA AVTQ |
-Macromolecule #3: Capsid protein VP3
| Macromolecule | Name: Capsid protein VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Possum enterovirus W6 Possum enterovirus W6 |
| Molecular weight | Theoretical: 26.997758 KDa |
| Sequence | String: GCPTLYTPGS GQFLTTDDFQ TPCMLPKFQP TPVIDIPGEV KNFLEVIQVE SLVEINNVSG VEGVARYRIP LNVQDAMDGQ IMAVRVDPG ADGPMQSTLL GVFTRYYTQW SGSLDFTFMF CGTFMTTGKV IIAYTPPGGD QPGSRQQAML GTHVVWDFGL Q SSITLVVP ...String: GCPTLYTPGS GQFLTTDDFQ TPCMLPKFQP TPVIDIPGEV KNFLEVIQVE SLVEINNVSG VEGVARYRIP LNVQDAMDGQ IMAVRVDPG ADGPMQSTLL GVFTRYYTQW SGSLDFTFMF CGTFMTTGKV IIAYTPPGGD QPGSRQQAML GTHVVWDFGL Q SSITLVVP WISSGHFRGT SLDNTIYKYR YYEAGYITMW YQTNMVVPPN FPTEASILMF VAAQPNFSLR ILKDRPDITQ VA SLQ |
-Macromolecule #4: Capsid protein VP4
| Macromolecule | Name: Capsid protein VP4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Possum enterovirus W6 Possum enterovirus W6 |
| Molecular weight | Theoretical: 7.63234 KDa |
| Sequence | String: MGAQLSKNTA GSHTTGTYAT GGSSIHYTNI NYYENAASNS LNKQDFTQDP DKFTRPVADI MKEAAVPLKS P |
-Macromolecule #5: SPHINGOSINE
| Macromolecule | Name: SPHINGOSINE / type: ligand / ID: 5 / Number of copies: 1 / Formula: SPH |
|---|---|
| Molecular weight | Theoretical: 299.492 Da |
| Chemical component information |  ChemComp-SPH: |
-Macromolecule #6: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 21 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.37 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.75 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)