+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-25819 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Lysophosphatidic acid receptor 1-Gi complex bound to LPA | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | GPCR / complex / lipid / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationAdenylate cyclase inhibitory pathway / cellular response to 1-oleoyl-sn-glycerol 3-phosphate / lysophosphatidic acid receptor activity / positive regulation of smooth muscle cell chemotaxis / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / Lysosphingolipid and LPA receptors / lysophosphatidic acid binding / Extra-nuclear estrogen signaling / Olfactory Signaling Pathway ...Adenylate cyclase inhibitory pathway / cellular response to 1-oleoyl-sn-glycerol 3-phosphate / lysophosphatidic acid receptor activity / positive regulation of smooth muscle cell chemotaxis / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / Lysosphingolipid and LPA receptors / lysophosphatidic acid binding / Extra-nuclear estrogen signaling / Olfactory Signaling Pathway / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / negative regulation of cilium assembly / corpus callosum development / G alpha (i) signalling events / bleb assembly / Activation of the phototransduction cascade / cellular response to oxygen levels / oligodendrocyte development / optic nerve development / regulation of synaptic vesicle cycle / regulation of metabolic process / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor / Presynaptic function of Kainate receptors / G beta:gamma signalling through CDC42 / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G alpha (12/13) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Ca2+ pathway / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (z) signalling events / Extra-nuclear estrogen signaling / G alpha (s) signalling events / G alpha (q) signalling events / G alpha (i) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / regulation of postsynaptic neurotransmitter receptor internalization / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Vasopressin regulates renal water homeostasis via Aquaporins / positive regulation of Rho protein signal transduction / positive regulation of dendritic spine development / G-protein alpha-subunit binding / negative regulation of cAMP/PKA signal transduction / adenylate cyclase inhibitor activity / positive regulation of stress fiber assembly / positive regulation of protein localization to cell cortex / T cell migration / response to prostaglandin E / D2 dopamine receptor binding / myelination / neurogenesis / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / cerebellum development / regulation of mitotic spindle organization / dendritic shaft / cell chemotaxis / PDZ domain binding / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / GABA-ergic synapse / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / adenylate cyclase-activating G protein-coupled receptor signaling pathway / photoreceptor disc membrane / GDP binding / cellular response to catecholamine stimulus / adenylate cyclase-activating dopamine receptor signaling pathway / regulation of cell shape / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / negative regulation of neuron projection development / sensory perception of taste / signaling receptor complex adaptor activity / positive regulation of cytosolic calcium ion concentration / retina development in camera-type eye / G protein activity / GTPase binding / presynaptic membrane / midbody / cell cortex / G alpha (i) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / dendritic spine Similarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.83 Å | ||||||||||||
 Authors Authors | Liu S / Paknejad N | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate. Authors: Shian Liu / Navid Paknejad / Lan Zhu / Yasuyuki Kihara / Manisha Ray / Jerold Chun / Wei Liu / Richard K Hite / Xin-Yun Huang /  Abstract: Lysophospholipids are bioactive lipids and can signal through G-protein-coupled receptors (GPCRs). The best studied lysophospholipids are lysophosphatidic acid (LPA) and sphingosine 1-phosphate (S1P). ...Lysophospholipids are bioactive lipids and can signal through G-protein-coupled receptors (GPCRs). The best studied lysophospholipids are lysophosphatidic acid (LPA) and sphingosine 1-phosphate (S1P). The mechanisms of lysophospholipid recognition by an active GPCR, and the activations of lysophospholipid GPCR-G-protein complexes remain unclear. Here we report single-particle cryo-EM structures of human S1P receptor 1 (S1P) and heterotrimeric G complexes formed with bound S1P or the multiple sclerosis (MS) treatment drug Siponimod, as well as human LPA receptor 1 (LPA) and G complexes in the presence of LPA. Our structural and functional data provide insights into how LPA and S1P adopt different conformations to interact with their cognate GPCRs, the selectivity of the homologous lipid GPCRs for S1P versus LPA, and the different activation mechanisms of these GPCRs by LPA and S1P. Our studies also reveal specific optimization strategies to improve the MS-treating S1P-targeting drugs. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25819.map.gz emd_25819.map.gz | 10.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25819-v30.xml emd-25819-v30.xml emd-25819.xml emd-25819.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25819.png emd_25819.png | 89.4 KB | ||
| Filedesc metadata |  emd-25819.cif.gz emd-25819.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25819 http://ftp.pdbj.org/pub/emdb/structures/EMD-25819 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25819 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25819 | HTTPS FTP |
-Related structure data
| Related structure data |  7td0MC  7td1C  7td2C  7td3C  7td4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25819.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25819.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.064 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : complex of Lysophosphatidic Acid Receptor 1 with G-protein and LPA
| Entire | Name: complex of Lysophosphatidic Acid Receptor 1 with G-protein and LPA |
|---|---|
| Components |
|
-Supramolecule #1: complex of Lysophosphatidic Acid Receptor 1 with G-protein and LPA
| Supramolecule | Name: complex of Lysophosphatidic Acid Receptor 1 with G-protein and LPA type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Lysophosphatidic acid receptor 1
| Macromolecule | Name: Lysophosphatidic acid receptor 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.734605 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDKAA AAAISTSIPV ISQPQFTAMN EPQCFYNESI AFFYNRSGKH LATEWNTVSK LVMGLGITVC IFIMLANLLV MVAIYVNRR FHFPIYYLMA NLAAADFFAG LAYFYLMFNT GPNTRRLTVS TWLLRQGLID TSLTASVANL LAIAIERHIT V FRMQLHTR ...String: DYKDDDDKAA AAAISTSIPV ISQPQFTAMN EPQCFYNESI AFFYNRSGKH LATEWNTVSK LVMGLGITVC IFIMLANLLV MVAIYVNRR FHFPIYYLMA NLAAADFFAG LAYFYLMFNT GPNTRRLTVS TWLLRQGLID TSLTASVANL LAIAIERHIT V FRMQLHTR MSNRRVVVVI VVIWTMAIVM GAIPSVGWNC ICDIENCSNM APLYSDSYLV FWAIFNLVTF VVMVVLYAHI FG YVRQRTM RMSRHSSGPR RNRDTMMSLL KTVVIVLGAF IICWTPGLVL LLLDVCCPQC DVLAYEKFFL LLAEFNSAMN PII YSYRDK EMSATFRQIL CCQRSENPTG PTEG UniProtKB: Lysophosphatidic acid receptor 1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.41693 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.16307 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLEVLFQG PHMASMGCTL SAEDKAAVER SKMIDRNLRE DGEKAAREVK LLLLGAGESG KSTIVKQMKI IHEAGYSEE ECKQYKAVVY SNTIQSIIAI IRAMGRLKID FGDSARADDA RQLFVLAGAA EEGFMTAELA GVIKRLWKDS G VQACFNRS ...String: MGSSHHHHHH SSGLEVLFQG PHMASMGCTL SAEDKAAVER SKMIDRNLRE DGEKAAREVK LLLLGAGESG KSTIVKQMKI IHEAGYSEE ECKQYKAVVY SNTIQSIIAI IRAMGRLKID FGDSARADDA RQLFVLAGAA EEGFMTAELA GVIKRLWKDS G VQACFNRS REYQLNDSAA YYLNDLDRIA QPNYIPTQQD VLRTRVKTTG IVETHFTFKD LHFKMFDVGA QRSERKKWIH CF EGVTAII FCVALSDYDL VLAEDEEMNR MHESMKLFDS ICNNKWFTDT SIILFLNKKD LFEEKIKKSP LTICYPEYAG SNT YEEAAA YIQCQFEDLN KRKDTKEIYT HFTCATDTKN VQFVFDAVTD VIIKNNLKDC GLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.845078 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFSAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate
| Macromolecule | Name: (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate type: ligand / ID: 5 / Number of copies: 1 / Formula: NKP |
|---|---|
| Molecular weight | Theoretical: 436.52 Da |
| Chemical component information | 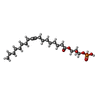 ChemComp-NKP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 28.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.83 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 1588791 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller


























 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)





















