[English] 日本語
 Yorodumi
Yorodumi- EMDB-24729: CryoEM structure of RBD domain of COVID-19 in complex with Legobody -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24729 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of RBD domain of COVID-19 in complex with Legobody | |||||||||
 Map data Map data | sharpened map for the complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RBD / maltose-binding protein / Fab / nanobody / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationIgG binding / carbohydrate transmembrane transporter activity / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion ...IgG binding / carbohydrate transmembrane transporter activity / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / periplasmic space / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / extracellular region / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Streptococcus sp. (bacteria) / Streptococcus sp. (bacteria) /    | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Wu XD / Rapoport TA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Cryo-EM structure determination of small proteins by nanobody-binding scaffolds (Legobodies). Authors: Xudong Wu / Tom A Rapoport /  Abstract: We describe a general method that allows structure determination of small proteins by single-particle cryo-electron microscopy (cryo-EM). The method is based on the availability of a target-binding ...We describe a general method that allows structure determination of small proteins by single-particle cryo-electron microscopy (cryo-EM). The method is based on the availability of a target-binding nanobody, which is then rigidly attached to two scaffolds: 1) a Fab fragment of an antibody directed against the nanobody and 2) a nanobody-binding protein A fragment fused to maltose binding protein and Fab-binding domains. The overall ensemble of ∼120 kDa, called Legobody, does not perturb the nanobody-target interaction, is easily recognizable in EM images due to its unique shape, and facilitates particle alignment in cryo-EM image processing. The utility of the method is demonstrated for the KDEL receptor, a 23-kDa membrane protein, resulting in a map at 3.2-Å overall resolution with density sufficient for de novo model building, and for the 22-kDa receptor-binding domain (RBD) of SARS-CoV-2 spike protein, resulting in a map at 3.6-Å resolution that allows analysis of the binding interface to the nanobody. The Legobody approach thus overcomes the current size limitations of cryo-EM analysis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24729.map.gz emd_24729.map.gz | 43.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24729-v30.xml emd-24729-v30.xml emd-24729.xml emd-24729.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24729.png emd_24729.png | 45.9 KB | ||
| Filedesc metadata |  emd-24729.cif.gz emd-24729.cif.gz | 6.9 KB | ||
| Others |  emd_24729_additional_1.map.gz emd_24729_additional_1.map.gz | 35.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24729 http://ftp.pdbj.org/pub/emdb/structures/EMD-24729 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24729 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24729 | HTTPS FTP |
-Related structure data
| Related structure data |  7rxdMC  7r9dC  7rxcC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24729.map.gz / Format: CCP4 / Size: 46.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24729.map.gz / Format: CCP4 / Size: 46.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map for the complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
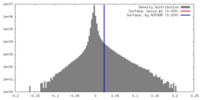
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unsharpened map
| File | emd_24729_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
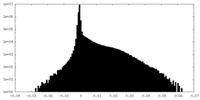
| Projections & Slices |
| ||||||||||||
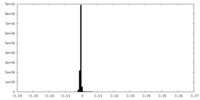
| Density Histograms |
- Sample components
Sample components
-Entire : The complex of RBD domain of COVID-19 with Legobody
| Entire | Name: The complex of RBD domain of COVID-19 with Legobody |
|---|---|
| Components |
|
-Supramolecule #1: The complex of RBD domain of COVID-19 with Legobody
| Supramolecule | Name: The complex of RBD domain of COVID-19 with Legobody / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: Maltodextrin-binding protein,Immunoglobulin G-binding protein A,I...
| Macromolecule | Name: Maltodextrin-binding protein,Immunoglobulin G-binding protein A,Immunoglobulin G-binding protein G type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptococcus sp. (bacteria) Streptococcus sp. (bacteria) |
| Molecular weight | Theoretical: 59.233246 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKIEEGKLVI WINGDKGYNG LAEVGKKFEK DTGIKVTVEH PDKLEEKFPQ VAATGDGPDI IFWAHDRFGG YAQSGLLAEI TPDKAFQDK LYPFTWDAVR YNGKLIAYPI AVEALSLIYN KDLLPNPPKT WEEIPALDKE LKAKGKSALM FNLQEPYFTW P LIAADGGY ...String: MKIEEGKLVI WINGDKGYNG LAEVGKKFEK DTGIKVTVEH PDKLEEKFPQ VAATGDGPDI IFWAHDRFGG YAQSGLLAEI TPDKAFQDK LYPFTWDAVR YNGKLIAYPI AVEALSLIYN KDLLPNPPKT WEEIPALDKE LKAKGKSALM FNLQEPYFTW P LIAADGGY AFKYENGKYD IKDVGVDNAG AKAGLTFLVD LIKNKHMNAD TDYSIAEAAF NKGETAMTIN GPWAWSNIDT SK VNYGVTV LPTFKGQPSK PFVGVLSAGI NAASPNKELA KEFLENYLLT DEGLEAVNKD KPLGAVALKS YEEELAKDPR IAA TMENAQ KGEIMPNIPQ MSAFWYAVRT AVINAASGRQ TVDQALAFAQ ILIMPNLTEE QRNGFIQSLK DDPSVSKEIL AEAK KLNEH QAPKGGSGGA GSGDQQSAFY EILNMPNLNE AQRNGFIQSL KDDPSQSTNV LGEAKKLNES QAGGGSGGGS GGSAV TTYK LVINGKTLKG ETTTKAVDAE TAEKAFKQYA NDNGVDGVWT YDDATKTFTV TEGSGHHHHH H UniProtKB: Maltodextrin-binding protein, Immunoglobulin G-binding protein A, Immunoglobulin G-binding protein G |
-Macromolecule #2: Fab_8D3_2 heavy chain
| Macromolecule | Name: Fab_8D3_2 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.252217 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DVQLVESGGG LVQPGKSLRL SCAASGFTFS NFGMHWVRQA PEMGLEWVAY ISSGSTTKYY GDTVKGRFTI SRDNPKNTLY LQMNSLRSE DTAMYYCARR PLYDGDYGYP MDYWGQGTSV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA ...String: DVQLVESGGG LVQPGKSLRL SCAASGFTFS NFGMHWVRQA PEMGLEWVAY ISSGSTTKYY GDTVKGRFTI SRDNPKNTLY LQMNSLRSE DTAMYYCARR PLYDGDYGYP MDYWGQGTSV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKVEPKSCGS HHHHHH |
-Macromolecule #3: Fab_8D3_2 light chain
| Macromolecule | Name: Fab_8D3_2 light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.095852 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NIMLTQSPSS LAVSAGERVT MSCKSTQSIL YNSNQKTYLA WYQQKPGQSP KLLIYWASTR ASGVPDRFTG SGSGTDFTLT INSVQPEDL AVYYCHQYLS AWTFGGGTKL EIKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV ...String: NIMLTQSPSS LAVSAGERVT MSCKSTQSIL YNSNQKTYLA WYQQKPGQSP KLLIYWASTR ASGVPDRFTG SGSGTDFTLT INSVQPEDL AVYYCHQYLS AWTFGGGTKL EIKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV TEQDSKDSTY SLSSTLTLSK ADYEKHKVYA CEVTHQGLSS PVTKSFNRGE C |
-Macromolecule #4: Nb_RBD
| Macromolecule | Name: Nb_RBD / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.661201 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLVESGGG LVQAGGSLRL SCAASGFPVY RDRMAWYRQA PGKEREWVAA IYSAGQQTRY ADSVKGRFTI SRDNAKNTVY LQMNSLKPE DTAVYYCNVK DVGHHYEYYD YWGQGTQVTV SSLEHHHHHH |
-Macromolecule #5: Spike protein S1
| Macromolecule | Name: Spike protein S1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.995555 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GDYKDDDDKG GENLYFQGGS GDSTGSSNLC PFGEVFNATR FASVYAWNRK RISNCVADYS VLYNSASFST FKCYGVSPTK LNDLCFTNV YADSFVIRGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYLYRL FRKSNLKPFE R DISTEIYQ ...String: GDYKDDDDKG GENLYFQGGS GDSTGSSNLC PFGEVFNATR FASVYAWNRK RISNCVADYS VLYNSASFST FKCYGVSPTK LNDLCFTNV YADSFVIRGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYLYRL FRKSNLKPFE R DISTEIYQ AGSTPCNGVE GFNCYFPLQS YGFQPTNGVG YQPYRVVVLS FELLHAPATV CGGGGSGSGH HHHHHHH UniProtKB: Spike glycoprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.42 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)