+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23938 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human SSU processome, state post-A1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosomal assembly intermediate / RIBOSOME | |||||||||
| Function / homology |  Function and homology information Function and homology informationU6 snRNA 2'-O-ribose methyltransferase activity / oocyte growth / nucleolar exosome (RNase complex) / nucleologenesis / 18S rRNA (adenine1779-N6/adenine1780-N6)-dimethyltransferase / 18S rRNA (adenine(1779)-N(6)/adenine(1780)-N(6))-dimethyltransferase activity / positive regulation of mRNA cis splicing, via spliceosome / snoRNA localization / nuclear polyadenylation-dependent antisense transcript catabolic process / nuclear polyadenylation-dependent snoRNA catabolic process ...U6 snRNA 2'-O-ribose methyltransferase activity / oocyte growth / nucleolar exosome (RNase complex) / nucleologenesis / 18S rRNA (adenine1779-N6/adenine1780-N6)-dimethyltransferase / 18S rRNA (adenine(1779)-N(6)/adenine(1780)-N(6))-dimethyltransferase activity / positive regulation of mRNA cis splicing, via spliceosome / snoRNA localization / nuclear polyadenylation-dependent antisense transcript catabolic process / nuclear polyadenylation-dependent snoRNA catabolic process / nuclear polyadenylation-dependent snRNA catabolic process / granular component / RNA exonuclease activity / nuclear polyadenylation-dependent CUT catabolic process / regulation of telomerase RNA localization to Cajal body / positive regulation of male gonad development / U4atac snRNP / TRAMP-dependent tRNA surveillance pathway / CURI complex / UTP-C complex / CUT catabolic process / regulation of stem cell population maintenance / exosome (RNase complex) / cytoplasmic exosome (RNase complex) / t-UTP complex / nuclear polyadenylation-dependent rRNA catabolic process / poly(A)-dependent snoRNA 3'-end processing / U4atac snRNA binding / Mpp10 complex / Pwp2p-containing subcomplex of 90S preribosome / nuclear exosome (RNase complex) / rRNA (pseudouridine) methyltransferase activity / exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / pre-snoRNP complex / box C/D sno(s)RNA binding / preribosome / histone H2AQ104 methyltransferase activity / rRNA (adenine-N6,N6-)-dimethyltransferase activity / histone mRNA catabolic process / box C/D sno(s)RNA 3'-end processing / endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / rRNA methyltransferase activity / endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / nuclear mRNA surveillance / dense fibrillar component / histone methyltransferase binding / endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / regulation of transcription elongation by RNA polymerase II / box C/D methylation guide snoRNP complex / positive regulation of rRNA processing / tRNA export from nucleus / cilium disassembly / rRNA primary transcript binding / transcription elongation factor activity / RNA splicing, via transesterification reactions / sno(s)RNA-containing ribonucleoprotein complex / spindle assembly involved in female meiosis / rRNA base methylation / blastocyst formation / U4 snRNA binding / positive regulation of respiratory burst involved in inflammatory response / protein localization to nucleolus / epigenetic programming in the zygotic pronuclei / Cul4-RING E3 ubiquitin ligase complex / SUMOylation of RNA binding proteins / nucleolus organization / U2-type precatalytic spliceosome / telomerase RNA binding / rRNA methylation / negative regulation of RNA splicing / RNA catabolic process / box C/D snoRNP assembly / positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator / neural precursor cell proliferation / neural crest cell differentiation / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / U3 snoRNA binding / negative regulation of bicellular tight junction assembly / rRNA modification in the nucleus and cytosol / Formation of the ternary complex, and subsequently, the 43S complex / erythrocyte homeostasis / cytoplasmic side of rough endoplasmic reticulum membrane / precatalytic spliceosome / snoRNA binding / preribosome, small subunit precursor / maturation of 5.8S rRNA / negative regulation of ubiquitin protein ligase activity / Ribosomal scanning and start codon recognition / Translation initiation complex formation / rRNA metabolic process / positive regulation of transcription by RNA polymerase I / negative regulation of telomere maintenance via telomerase / fibroblast growth factor binding / Association of TriC/CCT with target proteins during biosynthesis / monocyte chemotaxis / Protein hydroxylation / TOR signaling / RNA polymerase II complex binding / TFIID-class transcription factor complex binding / SARS-CoV-1 modulates host translation machinery Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
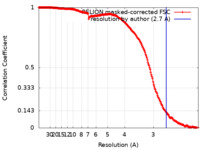
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Vanden Broeck A / Singh S / Klinge S | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Nucleolar maturation of the human small subunit processome. Authors: Sameer Singh / Arnaud Vanden Broeck / Linamarie Miller / Malik Chaker-Margot / Sebastian Klinge /  Abstract: The human small subunit processome mediates early maturation of the small ribosomal subunit by coupling RNA folding to subsequent RNA cleavage and processing steps. We report the high-resolution ...The human small subunit processome mediates early maturation of the small ribosomal subunit by coupling RNA folding to subsequent RNA cleavage and processing steps. We report the high-resolution cryo–electron microscopy structures of maturing human small subunit (SSU) processomes at resolutions of 2.7 to 3.9 angstroms. These structures reveal the molecular mechanisms that enable crucial progressions during SSU processome maturation. RNA folding states within these particles are communicated to and coordinated with key enzymes that drive irreversible steps such as targeted exosome-mediated RNA degradation, protein-guided site-specific endonucleolytic RNA cleavage, and tightly controlled RNA unwinding. These conserved mechanisms highlight the SSU processome’s impressive structural plasticity, which endows this 4.5-megadalton nucleolar assembly with the distinctive ability to mature the small ribosomal subunit from within. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23938.map.gz emd_23938.map.gz | 619.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23938-v30.xml emd-23938-v30.xml emd-23938.xml emd-23938.xml | 126.5 KB 126.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23938_fsc.xml emd_23938_fsc.xml | 19.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_23938.png emd_23938.png | 163.1 KB | ||
| Masks |  emd_23938_msk_1.map emd_23938_msk_1.map | 669.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23938.cif.gz emd-23938.cif.gz | 31.3 KB | ||
| Others |  emd_23938_additional_1.map.gz emd_23938_additional_1.map.gz emd_23938_half_map_1.map.gz emd_23938_half_map_1.map.gz emd_23938_half_map_2.map.gz emd_23938_half_map_2.map.gz | 616.4 MB 536.2 MB 536.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23938 http://ftp.pdbj.org/pub/emdb/structures/EMD-23938 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23938 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23938 | HTTPS FTP |
-Validation report
| Summary document |  emd_23938_validation.pdf.gz emd_23938_validation.pdf.gz | 1009.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23938_full_validation.pdf.gz emd_23938_full_validation.pdf.gz | 1009.4 KB | Display | |
| Data in XML |  emd_23938_validation.xml.gz emd_23938_validation.xml.gz | 27.8 KB | Display | |
| Data in CIF |  emd_23938_validation.cif.gz emd_23938_validation.cif.gz | 37.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23938 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23938 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23938 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23938 | HTTPS FTP |
-Related structure data
| Related structure data |  7mqaMC  7mq8C  7mq9C  7mqjC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10781 (Title: Nucleolar maturation of the human small subunit processome EMPIAR-10781 (Title: Nucleolar maturation of the human small subunit processomeData size: 74.6 TB Data #1: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 1 [micrographs - multiframe] Data #2: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 2 [micrographs - multiframe] Data #3: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 3 [micrographs - multiframe] Data #4: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 4 [micrographs - multiframe] Data #5: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 5 [micrographs - multiframe] Data #6: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 6 [micrographs - multiframe] Data #7: Aligned and averaged micrographs of human SSU processomes - Dataset 1 [micrographs - single frame] Data #8: Aligned and averaged micrographs of human SSU processomes - Dataset 2 [micrographs - single frame] Data #9: Aligned and averaged micrographs of human SSU processomes - Dataset 3 [micrographs - single frame] Data #10: Aligned and averaged micrographs of human SSU processomes - Dataset 4 [micrographs - single frame] Data #11: Aligned and averaged micrographs of human SSU processomes - Dataset 5 [micrographs - single frame] Data #12: Aligned and averaged micrographs of human SSU processomes - Dataset 6 [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23938.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23938.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23938_msk_1.map emd_23938_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_23938_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_23938_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_23938_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Human SSU processome
+Supramolecule #1: Human SSU processome
+Macromolecule #1: 5'ETS rRNA
+Macromolecule #2: 18S rRNA
+Macromolecule #3: U3 snoRNA
+Macromolecule #4: 40S ribosomal protein S18
+Macromolecule #5: 40S ribosomal protein S4, X isoform
+Macromolecule #6: 40S ribosomal protein S5
+Macromolecule #7: 40S ribosomal protein S6
+Macromolecule #8: 40S ribosomal protein S7
+Macromolecule #9: 40S ribosomal protein S8
+Macromolecule #10: 40S ribosomal protein S9
+Macromolecule #11: 40S ribosomal protein S12
+Macromolecule #12: 40S ribosomal protein S16
+Macromolecule #13: 40S ribosomal protein S11
+Macromolecule #14: 40S ribosomal protein S24
+Macromolecule #15: 40S ribosomal protein S28
+Macromolecule #16: WD repeat-containing protein 75
+Macromolecule #17: Nucleolar protein 11
+Macromolecule #18: U3 small nucleolar RNA-associated protein 15 homolog
+Macromolecule #19: WD repeat-containing protein 43
+Macromolecule #20: HEAT repeat-containing protein 1
+Macromolecule #21: U3 small nucleolar RNA-associated protein 4 homolog
+Macromolecule #22: Periodic tryptophan protein 2 homolog
+Macromolecule #23: U3 small nucleolar RNA-associated protein 6 homolog
+Macromolecule #24: WD repeat-containing protein 3
+Macromolecule #25: Transducin beta-like protein 3
+Macromolecule #26: U3 small nucleolar RNA-associated protein 18 homolog
+Macromolecule #27: WD repeat-containing protein 36
+Macromolecule #28: DDB1- and CUL4-associated factor 13
+Macromolecule #29: WD repeat-containing protein 46
+Macromolecule #30: Active regulator of SIRT1
+Macromolecule #31: U3 small nucleolar ribonucleoprotein protein IMP3
+Macromolecule #32: U3 small nucleolar ribonucleoprotein protein MPP10
+Macromolecule #33: Something about silencing protein 10
+Macromolecule #34: Nucleolar protein 7
+Macromolecule #35: 40S ribosomal protein S13
+Macromolecule #36: 40S ribosomal protein S14
+Macromolecule #37: Nucleolar protein 6
+Macromolecule #38: Ribosomal RNA-processing protein 7 homolog A
+Macromolecule #39: Probable dimethyladenosine transferase
+Macromolecule #40: 40S ribosomal protein S3a
+Macromolecule #41: 40S ribosomal protein S15a
+Macromolecule #42: 40S ribosomal protein S19
+Macromolecule #43: 40S ribosomal protein S27
+Macromolecule #44: RRP12-like protein
+Macromolecule #45: Probable ATP-dependent RNA helicase DHX37
+Macromolecule #46: Ubiquitin-40S ribosomal protein S27a
+Macromolecule #47: 40S ribosomal protein S17
+Macromolecule #48: Exosome component 10
+Macromolecule #49: Nucleolar protein 56
+Macromolecule #50: Nucleolar protein 58
+Macromolecule #51: rRNA 2'-O-methyltransferase fibrillarin
+Macromolecule #52: NHP2-like protein 1
+Macromolecule #53: U3 small nucleolar RNA-interacting protein 2
+Macromolecule #54: RNA 3'-terminal phosphate cyclase-like protein
+Macromolecule #55: Ribosome biogenesis protein BMS1 homolog
+Macromolecule #56: Ribosomal RNA small subunit methyltransferase NEP1
+Macromolecule #57: rRNA-processing protein FCF1 homolog
+Macromolecule #58: U3 small nucleolar ribonucleoprotein protein IMP4
+Macromolecule #59: Small subunit processome component 20 homolog
+Macromolecule #60: Deoxynucleotidyltransferase terminal-interacting protein 2
+Macromolecule #61: 40S ribosomal protein S23
+Macromolecule #62: U3 small nucleolar RNA-associated protein 14 homolog A
+Macromolecule #63: Nucleolar protein 14
+Macromolecule #64: Nucleolar complex protein 4 homolog
+Macromolecule #65: RNA-binding protein PNO1
+Macromolecule #66: Unassigned peptides
+Macromolecule #67: Probable U3 small nucleolar RNA-associated protein 11
+Macromolecule #68: Bystin
+Macromolecule #69: MAGNESIUM ION
+Macromolecule #70: ADENOSINE-5'-TRIPHOSPHATE
+Macromolecule #71: ZINC ION
+Macromolecule #72: GUANOSINE-5'-TRIPHOSPHATE
+Macromolecule #73: S-ADENOSYL-L-HOMOCYSTEINE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 84904 / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL | ||||||||||||||||||||||||
| Output model |  PDB-7mqa: |
 Movie
Movie Controller
Controller














































































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)