登録情報 データベース : EMDB / ID : EMD-23510タイトル NHEJ Long-range synaptic complex Pre-synaptic complex 複合体 : Long-range synaptic Complex of NHEJタンパク質・ペプチド : X-ray repair cross-complementing protein 6タンパク質・ペプチド : X-ray repair cross-complementing protein 5タンパク質・ペプチド : DNA-dependent protein kinase catalytic subunitタンパク質・ペプチド : Unknown peptideDNA : DNA (31-MER)DNA : DNA (30-MER)タンパク質・ペプチド : DNA repair protein XRCC4タンパク質・ペプチド : Non-homologous end-joining factor 1タンパク質・ペプチド : DNA ligase 4リガンド : ADENOSINE-5'-DIPHOSPHATE / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Homo sapiens (ヒト)手法 / / 解像度 : 4.6 Å He Y / Chen S 資金援助 Organization Grant number 国 National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) R01GM135651 National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) 5T32GM008382 National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) U24GM129547 National Institutes of Health/National Cancer Institute (NIH/NCI) P01CA092584 American Cancer Society IRG-15-173-21 National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) R01GM047251
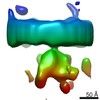
ジャーナル : Nature / 年 : 2021タイトル : Structural basis of long-range to short-range synaptic transition in NHEJ.著者 : Siyu Chen / Linda Lee / Tasmin Naila / Susan Fishbain / Annie Wang / Alan E Tomkinson / Susan P Lees-Miller / Yuan He / 要旨 : DNA double-strand breaks (DSBs) are a highly cytotoxic form of DNA damage and the incorrect repair of DSBs is linked to carcinogenesis. The conserved error-prone non-homologous end joining (NHEJ) ... DNA double-strand breaks (DSBs) are a highly cytotoxic form of DNA damage and the incorrect repair of DSBs is linked to carcinogenesis. The conserved error-prone non-homologous end joining (NHEJ) pathway has a key role in determining the effects of DSB-inducing agents that are used to treat cancer as well as the generation of the diversity in antibodies and T cell receptors. Here we applied single-particle cryo-electron microscopy to visualize two key DNA-protein complexes that are formed by human NHEJ factors. The Ku70/80 heterodimer (Ku), the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs), DNA ligase IV (LigIV), XRCC4 and XLF form a long-range synaptic complex, in which the DNA ends are held approximately 115 Å apart. Two DNA end-bound subcomplexes comprising Ku and DNA-PKcs are linked by interactions between the DNA-PKcs subunits and a scaffold comprising LigIV, XRCC4, XLF, XRCC4 and LigIV. The relative orientation of the DNA-PKcs molecules suggests a mechanism for autophosphorylation in trans, which leads to the dissociation of DNA-PKcs and the transition into the short-range synaptic complex. Within this complex, the Ku-bound DNA ends are aligned for processing and ligation by the XLF-anchored scaffold, and a single catalytic domain of LigIV is stably associated with a nick between the two Ku molecules, which suggests that the joining of both strands of a DSB involves both LigIV molecules. 履歴 登録 2021年2月18日 - ヘッダ(付随情報) 公開 2021年4月14日 - マップ公開 2021年4月14日 - 更新 2024年3月6日 - 現状 2024年3月6日 処理サイト : RCSB / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 マップデータ
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト) データ登録者
データ登録者 米国, 6件
米国, 6件  引用
引用 ジャーナル: Nature / 年: 2021
ジャーナル: Nature / 年: 2021

 構造の表示
構造の表示 ムービービューア
ムービービューア SurfView
SurfView Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク emd_23510.map.gz
emd_23510.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-23510-v30.xml
emd-23510-v30.xml emd-23510.xml
emd-23510.xml EMDBヘッダ
EMDBヘッダ emd_23510_fsc.xml
emd_23510_fsc.xml FSCデータファイル
FSCデータファイル emd_23510.png
emd_23510.png emd-23510.cif.gz
emd-23510.cif.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-23510
http://ftp.pdbj.org/pub/emdb/structures/EMD-23510 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23510
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23510 emd_23510_validation.pdf.gz
emd_23510_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_23510_full_validation.pdf.gz
emd_23510_full_validation.pdf.gz emd_23510_validation.xml.gz
emd_23510_validation.xml.gz emd_23510_validation.cif.gz
emd_23510_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23510
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23510 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23510
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23510 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_23510.map.gz / 形式: CCP4 / 大きさ: 93 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_23510.map.gz / 形式: CCP4 / 大きさ: 93 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) 試料の構成要素
試料の構成要素 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN 画像解析
画像解析
 ムービー
ムービー コントローラー
コントローラー






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















 unidentified baculovirus (ウイルス)
unidentified baculovirus (ウイルス)









