[English] 日本語
 Yorodumi
Yorodumi- EMDB-21924: Protofilament from a 13-protofilament, Taxol stabilized microtubule -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21924 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Protofilament from a 13-protofilament, Taxol stabilized microtubule | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Microtubule / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationMicrotubule-dependent trafficking of connexons from Golgi to the plasma membrane / Cilium Assembly / Intraflagellar transport / Carboxyterminal post-translational modifications of tubulin / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / Resolution of Sister Chromatid Cohesion / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / COPI-dependent Golgi-to-ER retrograde traffic ...Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Cilium Assembly / Intraflagellar transport / Carboxyterminal post-translational modifications of tubulin / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / Resolution of Sister Chromatid Cohesion / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / COPI-dependent Golgi-to-ER retrograde traffic / COPI-independent Golgi-to-ER retrograde traffic / COPI-mediated anterograde transport / RHO GTPases activate IQGAPs / RHO GTPases Activate Formins / MHC class II antigen presentation / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / Aggrephagy / The role of GTSE1 in G2/M progression after G2 checkpoint / Separation of Sister Chromatids / Recruitment of NuMA to mitotic centrosomes / Hedgehog 'off' state / positive regulation of axon guidance / intercellular bridge / microtubule-based process / structural constituent of cytoskeleton / cerebral cortex development / mitotic spindle / microtubule cytoskeleton organization / microtubule cytoskeleton / mitotic cell cycle / nervous system development / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / protein heterodimerization activity / GTPase activity / GTP binding / metal ion binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
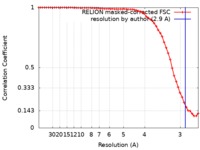
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Debs GE / Cha M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Dynamic and asymmetric fluctuations in the microtubule wall captured by high-resolution cryoelectron microscopy. Authors: Garrett E Debs / Michael Cha / Xueqi Liu / Andrew R Huehn / Charles V Sindelar /  Abstract: Microtubules are tubular polymers with essential roles in numerous cellular activities. Structures of microtubules have been captured at increasing resolution by cryo-EM. However, dynamic properties ...Microtubules are tubular polymers with essential roles in numerous cellular activities. Structures of microtubules have been captured at increasing resolution by cryo-EM. However, dynamic properties of the microtubule are key to its function, and this behavior has proved difficult to characterize at a structural level due to limitations in existing structure determination methods. We developed a high-resolution cryo-EM refinement method that divides an imaged microtubule into its constituent protofilaments, enabling deviations from helicity and other sources of heterogeneity to be quantified and corrected for at the single-subunit level. We demonstrate that this method improves the resolution of microtubule 3D reconstructions and substantially reduces anisotropic blurring artifacts, compared with methods that utilize helical symmetry averaging. Moreover, we identified an unexpected, discrete behavior of the m-loop, which mediates lateral interactions between neighboring protofilaments and acts as a flexible hinge between them. The hinge angle adopts preferred values corresponding to distinct conformations of the m-loop that are incompatible with helical symmetry. These hinge angles fluctuate in a stochastic manner, and perfectly cylindrical microtubule conformations are thus energetically and entropically penalized. The hinge angle can diverge further from helical symmetry at the microtubule seam, generating a subpopulation of highly distorted microtubules. However, the seam-distorted subpopulation disappears in the presence of Taxol, a microtubule stabilizing agent. These observations provide clues into the structural origins of microtubule flexibility and dynamics and highlight the role of structural polymorphism in defining microtubule behavior. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21924.map.gz emd_21924.map.gz | 14.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21924-v30.xml emd-21924-v30.xml emd-21924.xml emd-21924.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21924_fsc.xml emd_21924_fsc.xml | 5.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_21924.png emd_21924.png | 61.1 KB | ||
| Filedesc metadata |  emd-21924.cif.gz emd-21924.cif.gz | 6 KB | ||
| Others |  emd_21924_half_map_1.map.gz emd_21924_half_map_1.map.gz emd_21924_half_map_2.map.gz emd_21924_half_map_2.map.gz | 14.5 MB 14.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21924 http://ftp.pdbj.org/pub/emdb/structures/EMD-21924 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21924 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21924 | HTTPS FTP |
-Validation report
| Summary document |  emd_21924_validation.pdf.gz emd_21924_validation.pdf.gz | 902.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21924_full_validation.pdf.gz emd_21924_full_validation.pdf.gz | 902.2 KB | Display | |
| Data in XML |  emd_21924_validation.xml.gz emd_21924_validation.xml.gz | 11.3 KB | Display | |
| Data in CIF |  emd_21924_validation.cif.gz emd_21924_validation.cif.gz | 15.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21924 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21924 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21924 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21924 | HTTPS FTP |
-Related structure data
| Related structure data |  6wvrMC  6wvlC  6wvmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21924.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21924.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.333 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_21924_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_21924_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Microtubules stabilized with Taxol in the presence of a low conce...
| Entire | Name: Microtubules stabilized with Taxol in the presence of a low concentration of Kinesin Dimer and ADP |
|---|---|
| Components |
|
-Supramolecule #1: Microtubules stabilized with Taxol in the presence of a low conce...
| Supramolecule | Name: Microtubules stabilized with Taxol in the presence of a low concentration of Kinesin Dimer and ADP type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Tubulin alpha-1B chain
| Macromolecule | Name: Tubulin alpha-1B chain / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.204445 KDa |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE FSIYPAPQVS TAVVEPYNSI LTTHTTLEHS DCAFMVDNEA IYDICRRNLD IERPTYTNLN RLISQIVSSI TA SLRFDGA LNVDLTEFQT NLVPYPRIHF PLATYAPVIS AEKAYHEQLS VAEITNACFE PANQMVKCDP RHGKYMACCL LYR GDVVPK DVNAAIATIK TKRSIQFVDW CPTGFKVGIN YQPPTVVPGG DLAKVQRAVC MLSNTTAIAE AWARLDHKFD LMYA KRAFV HWYVGEGMEE GEFSEAREDM AALEKDYEEV GVDSVEGEGE EEGEEY UniProtKB: Tubulin alpha-1B chain |
-Macromolecule #2: Tubulin beta chain
| Macromolecule | Name: Tubulin beta chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.953797 KDa |
| Sequence | String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS ...String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS VMPSPKVSDT VVEPYNATLS VHQLVENTDE TYSIDNEALY DICFRTLKLT TPTYGDLNHL VSATMSGVTT CL RFPGQLN ADLRKLAVNM VPFPRLHFFM PGFAPLTSRG SQQYRALTVP ELTQQMFDSK NMMAACDPRH GRYLTVAAIF RGR MSMKEV DEQMLNVQNK NSSYFVEWIP NNVKTAVCDI PPRGLKMSAT FIGNSTAIQE LFKRISEQFT AMFRRKAFLH WYTG EGMDE MEFTEAESNM NDLVSEYQQY QDATADEQGE FEEEEGEDEA UniProtKB: Tubulin beta chain |
-Macromolecule #3: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 2 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 2 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Macromolecule #6: TAXOL
| Macromolecule | Name: TAXOL / type: ligand / ID: 6 / Number of copies: 2 / Formula: TA1 |
|---|---|
| Molecular weight | Theoretical: 853.906 Da |
| Chemical component information |  ChemComp-TA1: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 2-15 / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)