[English] 日本語
 Yorodumi
Yorodumi- EMDB-21647: Enterovirus D68 in complex with human monoclonal antibody EV68-159 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21647 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Enterovirus D68 in complex with human monoclonal antibody EV68-159 | |||||||||||||||
 Map data Map data | Enterovirus D68 in complex with human monoclonal antibody EV68-159 | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | virus / enterovirus / antibody / Structural Genomics / Center for Structural Genomics of Infectious Diseases / CSGID / VIRUS-IMMUNE SYSTEM complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell membrane / cysteine-type peptidase activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / helicase activity / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane ...host cell membrane / cysteine-type peptidase activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / helicase activity / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / viral capsid / nucleoside-triphosphate phosphatase / host cell / channel activity / monoatomic ion transmembrane transport / host cell cytoplasm / RNA helicase activity / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / symbiont-mediated suppression of host gene expression / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / symbiont entry into host cell / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Enterovirus D68 Enterovirus D68 | |||||||||||||||
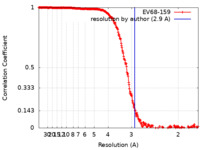
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||||||||
 Authors Authors | Fu J / Klose T | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Sci Immunol / Year: 2020 Journal: Sci Immunol / Year: 2020Title: Human antibodies neutralize enterovirus D68 and protect against infection and paralytic disease. Authors: Matthew R Vogt / Jianing Fu / Nurgun Kose / Lauren E Williamson / Robin Bombardi / Ian Setliff / Ivelin S Georgiev / Thomas Klose / Michael G Rossmann / Yury A Bochkov / James E Gern / ...Authors: Matthew R Vogt / Jianing Fu / Nurgun Kose / Lauren E Williamson / Robin Bombardi / Ian Setliff / Ivelin S Georgiev / Thomas Klose / Michael G Rossmann / Yury A Bochkov / James E Gern / Richard J Kuhn / James E Crowe /  Abstract: Enterovirus D68 (EV-D68) causes outbreaks of respiratory illness, and there is increasing evidence that it causes outbreaks of acute flaccid myelitis (AFM). There are no licensed therapies to prevent ...Enterovirus D68 (EV-D68) causes outbreaks of respiratory illness, and there is increasing evidence that it causes outbreaks of acute flaccid myelitis (AFM). There are no licensed therapies to prevent or treat EV-D68 infection or AFM disease. We isolated a panel of EV-D68-reactive human monoclonal antibodies that recognize diverse antigenic variants from participants with prior infection. One potently neutralizing cross-reactive antibody, EV68-228, protected mice from respiratory and neurologic disease when given either before or after infection. Cryo-electron microscopy studies revealed that EV68-228 and another potently neutralizing antibody (EV68-159) bound around the fivefold or threefold axes of symmetry on virion particles, respectively. The structures suggest diverse mechanisms of action by these antibodies. The high potency and effectiveness observed in vivo suggest that antibodies are a mechanistic correlate of protection against AFM disease and are candidates for clinical use in humans with EV-D68 infection. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21647.map.gz emd_21647.map.gz | 1.6 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21647-v30.xml emd-21647-v30.xml emd-21647.xml emd-21647.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21647_fsc.xml emd_21647_fsc.xml | 27 KB | Display |  FSC data file FSC data file |
| Images |  emd_21647.png emd_21647.png | 156.6 KB | ||
| Filedesc metadata |  emd-21647.cif.gz emd-21647.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21647 http://ftp.pdbj.org/pub/emdb/structures/EMD-21647 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21647 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21647 | HTTPS FTP |
-Related structure data
| Related structure data |  6wdsMC  6wdtC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21647.map.gz / Format: CCP4 / Size: 1.7 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21647.map.gz / Format: CCP4 / Size: 1.7 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Enterovirus D68 in complex with human monoclonal antibody EV68-159 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.874 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Enterovirus D68 in complex with human monoclonal antibody EV68-159
| Entire | Name: Enterovirus D68 in complex with human monoclonal antibody EV68-159 |
|---|---|
| Components |
|
-Supramolecule #1: Enterovirus D68 in complex with human monoclonal antibody EV68-159
| Supramolecule | Name: Enterovirus D68 in complex with human monoclonal antibody EV68-159 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #3: human antibody EV68-159
| Supramolecule | Name: human antibody EV68-159 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1, #6 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: Enterovirus D68
| Supramolecule | Name: Enterovirus D68 / type: virus / ID: 2 / Parent: 1 / Macromolecule list: #2-#5 / NCBI-ID: 42789 / Sci species name: Enterovirus D68 / Sci species strain: US/MO/14-18947 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: EV68-159 light chain
| Macromolecule | Name: EV68-159 light chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.715928 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QSVLTQPPSA SGTPGQRVTI SCSGSSSNIE YNYVYWYQKF PGTAPKLLIY KNNQRPSGVP DRFSGSKSGT SASLAISGLR SEDEGDYYC AAWDDILSGV VFGGGTKLTV L |
-Macromolecule #2: viral protein 1
| Macromolecule | Name: viral protein 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterovirus D68 / Strain: US/MO/14-18047 Enterovirus D68 / Strain: US/MO/14-18047 |
| Molecular weight | Theoretical: 32.920309 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: IESIIKTATD TVKSEINAEL GVVPSLNAVE TGATSNTEPE EAIQTRTVIN QHGVSETLVE NFLSRAALVS KRSFEYKDHT SSTARADKN FFKWTINTRS FVQLRRKLEL FTYLRFDAEI TILTTVAVNG SGNNTYVGLP DLTLQAMFVP TGALTPEKQD S FHWQSGSN ...String: IESIIKTATD TVKSEINAEL GVVPSLNAVE TGATSNTEPE EAIQTRTVIN QHGVSETLVE NFLSRAALVS KRSFEYKDHT SSTARADKN FFKWTINTRS FVQLRRKLEL FTYLRFDAEI TILTTVAVNG SGNNTYVGLP DLTLQAMFVP TGALTPEKQD S FHWQSGSN ASVFFKISDP PARITIPFMC INSAYSVFYD GFAGFEKNGL YGINPADTIG NLCVRIVNEH QPVGFTVTVR VY MKPKHIK AWAPRPPRTL PYMSIANANY KGKERAPNAL SAIIGNRDSV KTMPHNIVNT UniProtKB: Genome polyprotein |
-Macromolecule #3: viral protein 2
| Macromolecule | Name: viral protein 2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterovirus D68 / Strain: US/MO/14-18047 Enterovirus D68 / Strain: US/MO/14-18047 |
| Molecular weight | Theoretical: 27.567135 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SPSAEACGYS DRVLQLKLGN SAIVTQEAAN YCCAYGEWPN YLPDHEAVAI DKPTQPETAT DRFYTLKSVK WETGSTGWWW KLPDALNNI GMFGQNVQHH YLYRSGFLIH VQCNATKFHQ GALLVVAIPE HQRGAHNTNT SPGFDDIMKG EEGGTFNHPY V LDDGTSLA ...String: SPSAEACGYS DRVLQLKLGN SAIVTQEAAN YCCAYGEWPN YLPDHEAVAI DKPTQPETAT DRFYTLKSVK WETGSTGWWW KLPDALNNI GMFGQNVQHH YLYRSGFLIH VQCNATKFHQ GALLVVAIPE HQRGAHNTNT SPGFDDIMKG EEGGTFNHPY V LDDGTSLA CATIFPHQWI NLRTNNSATI VLPWMNAAPM DFPLRHNQWT LAIIPVVPLG TRTTSSMVPI TVSIAPMCCE FN GLRHAIT Q UniProtKB: Genome polyprotein |
-Macromolecule #4: viral protein 3
| Macromolecule | Name: viral protein 3 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterovirus D68 / Strain: US/MO/14-18047 Enterovirus D68 / Strain: US/MO/14-18047 |
| Molecular weight | Theoretical: 27.112814 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GVPTYLLPGS GQFLTTDDHS SAPALPCFNP TPEMHIPGQV RNMLEVVQVE SMMEINNTES AVGMERLKVD ISALTDVDQL LFNIPLDIQ LDGPLRNTLV GNISRYYTHW SGSLEMTFMF CGSFMAAGKL ILCYTPPGGS CPTTRETAML GTHIVWDFGL Q SSVTLIIP ...String: GVPTYLLPGS GQFLTTDDHS SAPALPCFNP TPEMHIPGQV RNMLEVVQVE SMMEINNTES AVGMERLKVD ISALTDVDQL LFNIPLDIQ LDGPLRNTLV GNISRYYTHW SGSLEMTFMF CGSFMAAGKL ILCYTPPGGS CPTTRETAML GTHIVWDFGL Q SSVTLIIP WISGSHYRMF NNDAKSTNAN VGYVTCFMQT NLIVPSESSD TCSLIGFIAA KDDFSLRLMR DSPDIGQLDH LH AAEAAYQ UniProtKB: Genome polyprotein |
-Macromolecule #5: viral protein 4
| Macromolecule | Name: viral protein 4 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterovirus D68 / Strain: US/MO/14-18047 Enterovirus D68 / Strain: US/MO/14-18047 |
| Molecular weight | Theoretical: 7.33696 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GAQVTRQQTG THENANIATN GSHITYNQIN FYKDSYAASA SKQDFSQDPS KFTEPVVEGL KAGAPVLK UniProtKB: Genome polyprotein |
-Macromolecule #6: EV68-159 heavy chain
| Macromolecule | Name: EV68-159 heavy chain / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.003529 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVKPGGLRLS CAASGFTFST YIMTWVRQAP GRGLEWVSSI STSSVYTFYA DSLKGRFTIS RDNAKNSVYL QMNSLRADD TAVYYCAREE GFRAYNLYWG QGTLVTVSS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 732 / Average exposure time: 12.0 sec. / Average electron dose: 31.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)