[English] 日本語
 Yorodumi
Yorodumi- EMDB-20693: Adeno-associated virus strain AAVhu.37 capsid icosahedral structure -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20693 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Adeno-associated virus strain AAVhu.37 capsid icosahedral structure | |||||||||
 Map data Map data | icosahedrally-averaged map of AAVhu.37 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | capsid / blood-brain barrier penetration / VIRUS | |||||||||
| Function / homology | Phospholipase A2-like domain / Phospholipase A2-like domain / Parvovirus coat protein VP2 / Parvovirus coat protein VP1/VP2 / Parvovirus coat protein VP1/VP2 / Capsid/spike protein, ssDNA virus / T=1 icosahedral viral capsid / structural molecule activity / Capsid protein VP1 Function and homology information Function and homology information | |||||||||
| Biological species |   Adeno-associated virus Adeno-associated virus | |||||||||
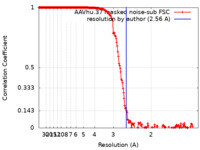
| Method | single particle reconstruction / cryo EM / Resolution: 2.56 Å | |||||||||
 Authors Authors | Kaelber JT / Yost SA | |||||||||
 Citation Citation |  Journal: Acta Crystallogr F Struct Biol Commun / Year: 2020 Journal: Acta Crystallogr F Struct Biol Commun / Year: 2020Title: Structure of the AAVhu.37 capsid by cryoelectron microscopy. Authors: Jason T Kaelber / Samantha A Yost / Keith A Webber / Emre Firlar / Ye Liu / Olivier Danos / Andrew C Mercer /  Abstract: Adeno-associated viruses (AAVs) are used as in vivo gene-delivery vectors in gene-therapy products and have been heavily investigated for numerous indications. Over 100 naturally occurring AAV ...Adeno-associated viruses (AAVs) are used as in vivo gene-delivery vectors in gene-therapy products and have been heavily investigated for numerous indications. Over 100 naturally occurring AAV serotypes and variants have been isolated from primate samples. Many reports have described unique properties of these variants (for instance, differences in potency, target cell or evasion of the immune response), despite high amino-acid sequence conservation. AAVhu.37 is of interest for clinical applications owing to its proficient transduction of the liver and central nervous system. The sequence identity of the AAVhu.37 VP1 to the well characterized AAVrh.10 serotype, for which no structure is available, is greater than 98%. Here, the structure of the AAVhu.37 capsid at 2.56 Å resolution obtained via single-particle cryo-electron microscopy is presented. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20693.map.gz emd_20693.map.gz | 609.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20693-v30.xml emd-20693-v30.xml emd-20693.xml emd-20693.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20693_fsc.xml emd_20693_fsc.xml | 21.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_20693.png emd_20693.png | 229 KB | ||
| Filedesc metadata |  emd-20693.cif.gz emd-20693.cif.gz | 6.6 KB | ||
| Others |  emd_20693_half_map_1.map.gz emd_20693_half_map_1.map.gz emd_20693_half_map_2.map.gz emd_20693_half_map_2.map.gz | 615.8 MB 615.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20693 http://ftp.pdbj.org/pub/emdb/structures/EMD-20693 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20693 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20693 | HTTPS FTP |
-Related structure data
| Related structure data |  6u95MC  7jotC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20693.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20693.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | icosahedrally-averaged map of AAVhu.37 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.70065 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: half A
| File | emd_20693_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half A | ||||||||||||
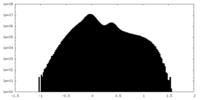
| Projections & Slices |
| ||||||||||||
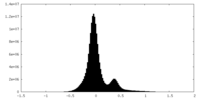
| Density Histograms |
-Half map: half B
| File | emd_20693_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half B | ||||||||||||
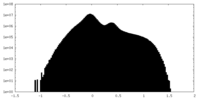
| Projections & Slices |
| ||||||||||||
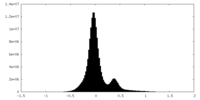
| Density Histograms |
- Sample components
Sample components
-Entire : Adeno-associated virus
| Entire | Name:   Adeno-associated virus Adeno-associated virus |
|---|---|
| Components |
|
-Supramolecule #1: Adeno-associated virus
| Supramolecule | Name: Adeno-associated virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Virions containing GFP-encoding ssDNA were assembled in transfected HEK293T helper cells. NCBI-ID: 272636 / Sci species name: Adeno-associated virus / Sci species strain: hu.37 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 5 MDa |
| Virus shell | Shell ID: 1 / Diameter: 250.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Adeno-associated virus Adeno-associated virus |
| Molecular weight | Theoretical: 81.528852 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AADGYLPDWL EDNLSEGIRE WWDLKPGAPK PKANQQKQDD GRGLVLPGYK YLGPFNGLDK GEPVNAADAA ALEHDKAYDQ QLKAGDNPY LRYNHADAEF QERLQEDTSF GGNLGRAVFQ AKKRVLEPLG LVEEAAKTAP GKKRPVEPSP QRSPDSSTGI G KKGQQPAK ...String: AADGYLPDWL EDNLSEGIRE WWDLKPGAPK PKANQQKQDD GRGLVLPGYK YLGPFNGLDK GEPVNAADAA ALEHDKAYDQ QLKAGDNPY LRYNHADAEF QERLQEDTSF GGNLGRAVFQ AKKRVLEPLG LVEEAAKTAP GKKRPVEPSP QRSPDSSTGI G KKGQQPAK KRLNFGQTGD SESVPDPQPI GEPPAGPSGL GSGTMAAGGG APMADNNEGA DGVGSSSGNW HCDSTWLGDR VI TTSTRTW ALPTYNNHLY KQISNGTSGG STNDNTYFGY STPWGYFDFN RFHCHFSPRD WQRLINNNWG FRPKRLSFKL FNI QVKEVT QNEGTKTIAN NLTSTIQVFT DSEYQLPYVL GSAHQGCLPP FPADVFMIPQ YGYLTLNNGS QAVGRSSFYC LEYF PSQML RTGNNFEFSY TFEDVPFHSS YAHSQSLDRL MNPLIDQYLY YLSRTQSTGG TQGTQQLLFS QAGPANMSAQ AKNWL PGPC YRQQRVSTTL SQNNNSNFAW TGATKYHLNG RDSLVNPGVA MATHKDDEER FFPSSGVLMF GKQGAGRDNV DYSSVM LTS EEEIKTTNPV ATEQYGVVAD NLQQTNTGPI VGNVNSQGAL PGMVWQNRDV YLQGPIWAKI PHTDGNFHPS PLMGGFG LK HPPPQILIKN TPVPADPPTT FSQAKLASFI TQYSTGQVSV EIEWELQKEN SKRWNPEIQY TSNYYKSTNV DFAVNTEG T YSEPRPIGTR YLTRNL UniProtKB: Capsid protein VP1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
Details: PBS | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2.86 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.037 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: LEICA EM GP Details: Whatman #1 filter paper, 4.5s blot. 3.5 microliters specimen applied.. | ||||||||||||
| Details | approx. 7*10^13 genome copies per mL |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Chromatic aberration corrector: none / Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-25 / Number grids imaged: 1 / Average exposure time: 5.0 sec. / Average electron dose: 1.31 e/Å2 / Details: collected in superresolution mode |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 1.584 µm / Calibrated defocus min: 0.535 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)