[English] 日本語
 Yorodumi
Yorodumi- EMDB-17588: Single particle cryo-EM of the P140-P110 heterodimer with an alte... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single particle cryo-EM of the P140-P110 heterodimer with an alternative conformation in the P140 stalk of Mycoplasma genitalium at a resolution of 3.7 Angstrom. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Adhesion / Mycoplasma genitalium / CELL ADHESION | |||||||||
| Function / homology |  Function and homology information Function and homology informationadhesion of symbiont to microvasculature / cell adhesion / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Mycoplasmoides genitalium G37 (bacteria) Mycoplasmoides genitalium G37 (bacteria) | |||||||||
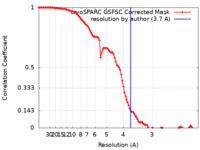
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Sprankel L / Scheffer MP / Frangakis AS | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2023 Journal: PLoS Pathog / Year: 2023Title: Cryo-electron tomography reveals the binding and release states of the major adhesion complex from Mycoplasma genitalium. Authors: Lasse Sprankel / Margot P Scheffer / Sina Manger / Utz H Ermel / Achilleas S Frangakis /  Abstract: The nap particle is an immunogenic surface adhesion complex from Mycoplasma genitalium. It is essential for motility and responsible for binding sialylated oligosaccharides on the surface of the host ...The nap particle is an immunogenic surface adhesion complex from Mycoplasma genitalium. It is essential for motility and responsible for binding sialylated oligosaccharides on the surface of the host cell. The nap particle is composed of two P140-P110 heterodimers, the structure of which was recently solved. However, the interpretation of the mechanism by which the mycoplasma cells orchestrate adhesion remained challenging. Here, we provide cryo-electron tomography structures at ~11 Å resolution, which allow for the distinction between the bound and released state of the nap particle, displaying the in vivo conformational states. Fitting of the atomically resolved structures reveals that bound sialylated oligosaccharides are stabilized by both P110 and P140. Movement of the stalk domains allows for the transfer of conformational changes from the interior of the cell to the binding pocket, thus having the capability of an active release process. It is likely that the same mechanism can be transferred to other Mycoplasma species that belong to the pneumoniae cluster. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17588.map.gz emd_17588.map.gz | 97.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17588-v30.xml emd-17588-v30.xml emd-17588.xml emd-17588.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17588_fsc.xml emd_17588_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_17588.png emd_17588.png | 187.9 KB | ||
| Masks |  emd_17588_msk_1.map emd_17588_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17588.cif.gz emd-17588.cif.gz | 7.5 KB | ||
| Others |  emd_17588_half_map_1.map.gz emd_17588_half_map_1.map.gz emd_17588_half_map_2.map.gz emd_17588_half_map_2.map.gz | 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17588 http://ftp.pdbj.org/pub/emdb/structures/EMD-17588 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17588 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17588 | HTTPS FTP |
-Related structure data
| Related structure data |  8pbyMC  8pbxC  8pbzC  8pc0C  8pc1C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17588.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17588.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
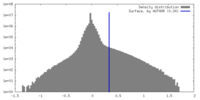
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17588_msk_1.map emd_17588_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17588_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17588_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : P140-P110 Heterodimer
| Entire | Name: P140-P110 Heterodimer |
|---|---|
| Components |
|
-Supramolecule #1: P140-P110 Heterodimer
| Supramolecule | Name: P140-P110 Heterodimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Mgp-operon protein 3
| Supramolecule | Name: Mgp-operon protein 3 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Mycoplasmoides genitalium G37 (bacteria) Mycoplasmoides genitalium G37 (bacteria) |
-Supramolecule #3: Adhesin P1
| Supramolecule | Name: Adhesin P1 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Mycoplasmoides genitalium G37 (bacteria) Mycoplasmoides genitalium G37 (bacteria) |
-Macromolecule #1: Mgp-operon protein 3
| Macromolecule | Name: Mgp-operon protein 3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycoplasmoides genitalium G37 (bacteria) Mycoplasmoides genitalium G37 (bacteria) |
| Molecular weight | Theoretical: 115.382062 KDa |
| Recombinant expression | Organism:  Mycoplasmoides genitalium G37 (bacteria) Mycoplasmoides genitalium G37 (bacteria) |
| Sequence | String: MKTMRKQIYK KAYWLLLPFL PLALANTFLV KEDSKNVTAY TPFATPITDS KSDLVSLAQL DSSYQIADQT IHNTNLFVLF KSRDVKVKY ESSGSNNISF DSTSQGEKPS YVVEFTNSTN IGIKWTMVKK YQLDVPNVSS DMNQVLKNLI LEQPLTKYTL N SSLAKEKG ...String: MKTMRKQIYK KAYWLLLPFL PLALANTFLV KEDSKNVTAY TPFATPITDS KSDLVSLAQL DSSYQIADQT IHNTNLFVLF KSRDVKVKY ESSGSNNISF DSTSQGEKPS YVVEFTNSTN IGIKWTMVKK YQLDVPNVSS DMNQVLKNLI LEQPLTKYTL N SSLAKEKG KTQREVHLGS GQANQWTSQR NQHDLNNNPS PNASTGFKLT TGNAYRKLSE SWPIYEPIDG TKQGKGKDSS GW SSTEENE AKNDAPSVSG GGSSSGTFNK YLNTKQALES IGILFDDQTP RNVITQLYYA STSKLAVTNN HIVVMGNSFL PSM WYWVVE RSAQENASNK PTWFANTNLD WGEDKQKQFV ENQLGYKETT STNSHNFHSK SFTQPAYLIS GIDSVNDQII FSGF KAGSV GYDSSSSSSS SSSSSTKDQA LAWSTTTSLD SKTGYKDLVT NDTGLNGPIN GSFSIQDTFS FVVPYSGNHT NNGTT GPIK TAYPVKKDQK STVKINSLIN ATPLNSYGDE GIGVFDALGL NYNFKSNQER LPSRTDQIFV YGIVSPNELR SAKSSA DST GSDTKVNWSN TQSRYLPVPY NYSEGIIDAD GFKRPENRGA SVTTFSGLKS IAPDGFANSI ANFSVGLKAG IDPNPVM SG KKANYGAVVL TRGGVVRLNF NPGNDSLLST TDNNIAPISF SFTPFTAAES AVDLTTFKEV TYNQESGLWS YIFDSSLK P SHDGKQTPVT DNMGFSVITV SRTGIELNQD QATTTLDVAP SALAVQSGIQ STTQTLTGVL PLSEEFSAVI AKDSDQNKI DIYKNNNGLF EIDTQLSNSV ATNNGGLAPS YTENRVDAWG KVEFADNSVL QARNLVDKTV DEIINTPEIL NSFFRFTPAF EDQKATLVA TKQSDTSLSV SPRIQFLDGN FYDLNSTIAG VPLNIGFPSR VFAGFAALPA WVIPVSVGSS VGILFILLVL G LGIGIPMY RVRKLQDASF VNVFKKVDTL TTAVGSVYKK IITQTGVVKK APSALKAANP SVKKPAAFLK PPVQPPSKPE GE QKAVEVK SEETKSHHHH HH UniProtKB: Mgp-operon protein 3 |
-Macromolecule #2: Adhesin P1
| Macromolecule | Name: Adhesin P1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycoplasmoides genitalium G37 (bacteria) Mycoplasmoides genitalium G37 (bacteria) |
| Molecular weight | Theoretical: 159.809297 KDa |
| Sequence | String: MHQPKKRLAK KSWAFLTAAL TLGVITGVGG YFLFNQNKQR SSVSNFAYQP KQLSVKHQQA VDETLTPWTW NNNNFSSLKI TGENPGSFG LVRSQNDNLN ISSVTKNSSD DNLKYLNAVE KYLDGQQNFA IRRYDNNGRA LYDINLAKME NPSTVQRGLN G EPIFDPFK ...String: MHQPKKRLAK KSWAFLTAAL TLGVITGVGG YFLFNQNKQR SSVSNFAYQP KQLSVKHQQA VDETLTPWTW NNNNFSSLKI TGENPGSFG LVRSQNDNLN ISSVTKNSSD DNLKYLNAVE KYLDGQQNFA IRRYDNNGRA LYDINLAKME NPSTVQRGLN G EPIFDPFK GFGLTGNAPT DWNEIKGKVP VEVVQSPHSP NLYFVLLVPK VALEYHNLNN QVVKESLEVK ATQSSFNPTQ RL QKDSPVK DSSKQGEKLS ETTASSMSSG MATSTRAKAL KVEVERGSQS DSLLKNDFAK KPLKHKNSSG EVKLEAEKEF TEA WKPLLT TDQIAREKGM GATVVSFYDA PYSENHTAFG LVDHIDPKKM VENYPPSWKT PKWNHHGIWD YNARNLLLQT TGFF NPRRH PEWFDEGQAK ADNTSPGFKV GDTDHKKDGF KKNSSSPIAL PFEAYFANIG NMVAIGNSVF IFGGNGHATK MFTTN PLSI GVFRIKYTDN FSKSSVTGWP YAVLFGGLIN PQTNGLKDLP LGTNRWFEYV PRMAVSGVKW VGNQLVLAGT LTMGDT ATV PRLKYDQLEK HLNLVAQGQG LLREDLQIFT PYGWANRPDI PVGAWLQDEM GSKFGPHYFL NNPDIQDNVN NDTVEAL IS SYKNTDKLKH VYPYRYSGLY AWQLFNWSNK LTNTPLSANF VNENSYAPNS LFAAILNEDL LTGLSDKIFY GKENEFAE N EADRFNQLLS LNPNPNTNWA RYLNVVQRFT TGPNLDSSTF DQFLDFLPWI GNGKPFSNSP SPSTSASSST PLPTFSNIN VGVKSMITQH LNKENTRWVF IPNFSPDIWT GAGYRVQSAN QKNGIPFEQV KPSNNSTPFD PNSDDNKVTP SGGSSKPTTY PALPNSISP TSDWINALTF TNKNNPQRNQ LLLRSLLGTI PVLINKSGDS NDQFNKDSEQ KWDKTETNEG NLPGFGEVNG L YNAALLHT YGFFGTNTNS TDPKIGFKAD SSSSSSSTLV GSGLNWTSQD VGNLVVINDT SFGFQLGGWF ITFTDFIRPR TG YLGITLS SLQDQTIIWA DQPWTSFKGS YLDSDGTPKS LWDPTALKSL PNSSTTYDTN PTLSPSFQLY QPNKVKAYQT TNT YNKLIE PVDATSAATN MTSLLKLLTT KNIKAKLGKG TASSQGNNNG GGVSQTINTI TTTGNISEGL KEETSIQAET LKKF FDSKQ NNKSEIGIGD STFTKMDGKL TGVVSTPLVN LINGQGATSD SDTEKISFKP GNQIDFNRLF TLPVTELFDP NTMFV YDQY VPLLVNLPSG FDQASIRLKV ISYSVENQTL GVRLEFKDPQ TQQFIPVLNA SSTGPQTVFQ PFNQWADYVL PLIVTV PIV VIILSVTLGL TIGIPMHRNK KALQAGFDLS NKKVDVLTKA VGSVFKEIIN RTGISNAPKK LKQATPTKPT PKTPPKP PV KQ UniProtKB: Adhesin P1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.025 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum SE / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 4.0 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 130000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL | ||||||
| Output model |  PDB-8pby: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)