[English] 日本語
 Yorodumi
Yorodumi- EMDB-17589: Single particle cryo-EM of the Nap complex of Mycoplasma genitali... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
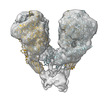
| Title | Single particle cryo-EM of the Nap complex of Mycoplasma genitalium in the expanded conformation at 7.3 Angstrom resolution. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Adhesion / Mycoplasma genitalium / CELL ADHESION | |||||||||
| Biological species |  Mycoplasmoides genitalium G37 (bacteria) Mycoplasmoides genitalium G37 (bacteria) | |||||||||
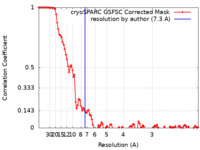
| Method | single particle reconstruction / cryo EM / Resolution: 7.3 Å | |||||||||
 Authors Authors | Sprankel L / Scheffer MP / Frangakis AS | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2023 Journal: PLoS Pathog / Year: 2023Title: Cryo-electron tomography reveals the binding and release states of the major adhesion complex from Mycoplasma genitalium. Authors: Lasse Sprankel / Margot P Scheffer / Sina Manger / Utz H Ermel / Achilleas S Frangakis /  Abstract: The nap particle is an immunogenic surface adhesion complex from Mycoplasma genitalium. It is essential for motility and responsible for binding sialylated oligosaccharides on the surface of the host ...The nap particle is an immunogenic surface adhesion complex from Mycoplasma genitalium. It is essential for motility and responsible for binding sialylated oligosaccharides on the surface of the host cell. The nap particle is composed of two P140-P110 heterodimers, the structure of which was recently solved. However, the interpretation of the mechanism by which the mycoplasma cells orchestrate adhesion remained challenging. Here, we provide cryo-electron tomography structures at ~11 Å resolution, which allow for the distinction between the bound and released state of the nap particle, displaying the in vivo conformational states. Fitting of the atomically resolved structures reveals that bound sialylated oligosaccharides are stabilized by both P110 and P140. Movement of the stalk domains allows for the transfer of conformational changes from the interior of the cell to the binding pocket, thus having the capability of an active release process. It is likely that the same mechanism can be transferred to other Mycoplasma species that belong to the pneumoniae cluster. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17589.map.gz emd_17589.map.gz | 96.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17589-v30.xml emd-17589-v30.xml emd-17589.xml emd-17589.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17589_fsc.xml emd_17589_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_17589.png emd_17589.png | 119.4 KB | ||
| Masks |  emd_17589_msk_1.map emd_17589_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17589.cif.gz emd-17589.cif.gz | 4.2 KB | ||
| Others |  emd_17589_half_map_1.map.gz emd_17589_half_map_1.map.gz emd_17589_half_map_2.map.gz emd_17589_half_map_2.map.gz | 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17589 http://ftp.pdbj.org/pub/emdb/structures/EMD-17589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17589 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17589.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17589.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17589_msk_1.map emd_17589_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
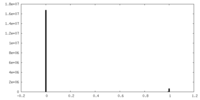
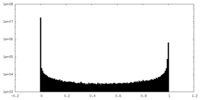
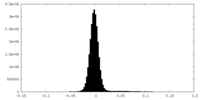
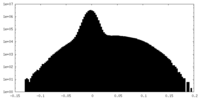
| Density Histograms |
-Half map: #1
| File | emd_17589_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
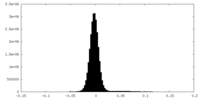
| Density Histograms |
-Half map: #2
| File | emd_17589_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
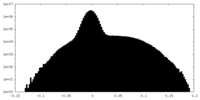
| Density Histograms |
- Sample components
Sample components
-Entire : Nap adhesion complex in the expanded conformation
| Entire | Name: Nap adhesion complex in the expanded conformation |
|---|---|
| Components |
|
-Supramolecule #1: Nap adhesion complex in the expanded conformation
| Supramolecule | Name: Nap adhesion complex in the expanded conformation / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Mycoplasmoides genitalium G37 (bacteria) Mycoplasmoides genitalium G37 (bacteria) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.025 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum SE / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 4.0 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 130000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)