[English] 日本語
 Yorodumi
Yorodumi- EMDB-12805: Resting state GluA1/A2 AMPA receptor in complex with TARP gamma 8... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12805 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Resting state GluA1/A2 AMPA receptor in complex with TARP gamma 8 and CNIH2 (LBD-TMD) | |||||||||
 Map data Map data | Composite map generated based on masked refinement maps of LBD and TMD. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AMPAR / ion channels / neurotransmission / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of receptor localization to synapse / negative regulation of anterograde synaptic vesicle transport / Phase 0 - rapid depolarisation / Phase 2 - plateau phase / Cargo concentration in the ER / axonal spine / positive regulation of locomotion involved in locomotory behavior / positive regulation of membrane potential / COPII-mediated vesicle transport / localization within membrane ...negative regulation of receptor localization to synapse / negative regulation of anterograde synaptic vesicle transport / Phase 0 - rapid depolarisation / Phase 2 - plateau phase / Cargo concentration in the ER / axonal spine / positive regulation of locomotion involved in locomotory behavior / positive regulation of membrane potential / COPII-mediated vesicle transport / localization within membrane / cellular response to ammonium ion / response to sucrose / L-type voltage-gated calcium channel complex / neuron spine / myosin V binding / postsynaptic neurotransmitter receptor diffusion trapping / proximal dendrite / channel regulator activity / regulation of monoatomic ion transmembrane transport / LGI-ADAM interactions / protein phosphatase 2B binding / Trafficking of AMPA receptors / regulation of AMPA receptor activity / response to arsenic-containing substance / cellular response to L-glutamate / cellular response to dsRNA / regulation of NMDA receptor activity / dendritic spine membrane / long-term synaptic depression / beta-2 adrenergic receptor binding / Synaptic adhesion-like molecules / cellular response to peptide hormone stimulus / spine synapse / dendritic spine neck / dendritic spine cytoplasm / neuronal cell body membrane / cellular response to amine stimulus / dendritic spine head / response to psychosocial stress / peptide hormone receptor binding / response to morphine / spinal cord development / perisynaptic space / Activation of AMPA receptors / ligand-gated monoatomic cation channel activity / protein kinase A binding / AMPA glutamate receptor activity / Trafficking of GluR2-containing AMPA receptors / response to lithium ion / behavioral response to pain / transmission of nerve impulse / AMPA glutamate receptor clustering / adenylate cyclase binding / cellular response to glycine / kainate selective glutamate receptor activity / AMPA glutamate receptor complex / immunoglobulin binding / asymmetric synapse / response to electrical stimulus / regulation of receptor recycling / extracellularly glutamate-gated ion channel activity / ionotropic glutamate receptor complex / G-protein alpha-subunit binding / conditioned place preference / Unblocking of NMDA receptors, glutamate binding and activation / glutamate receptor binding / long-term memory / positive regulation of synaptic transmission / regulation of synaptic transmission, glutamatergic / positive regulation of synaptic transmission, glutamatergic / regulation of postsynaptic membrane neurotransmitter receptor levels / postsynaptic density, intracellular component / neuronal action potential / response to fungicide / voltage-gated calcium channel activity / cytoskeletal protein binding / vesicle-mediated transport / extracellular ligand-gated monoatomic ion channel activity / glutamate-gated receptor activity / cellular response to brain-derived neurotrophic factor stimulus / regulation of long-term synaptic depression / synapse assembly / somatodendritic compartment / glutamate-gated calcium ion channel activity / presynaptic active zone membrane / dendrite membrane / excitatory synapse / ionotropic glutamate receptor binding / ionotropic glutamate receptor signaling pathway / dendrite cytoplasm / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / synaptic membrane / positive regulation of excitatory postsynaptic potential / dendritic shaft / SNARE binding / response to cocaine / PDZ domain binding / calcium channel regulator activity / neuromuscular junction / synaptic transmission, glutamatergic Similarity search - Function | |||||||||
| Biological species |  | |||||||||
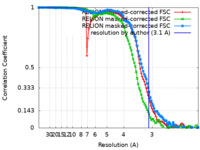
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Zhang D / Watson JF / Matthews PM / Cais O / Greger IH | |||||||||
| Funding support | 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Gating and modulation of a hetero-octameric AMPA glutamate receptor. Authors: Danyang Zhang / Jake F Watson / Peter M Matthews / Ondrej Cais / Ingo H Greger /   Abstract: AMPA receptors (AMPARs) mediate the majority of excitatory transmission in the brain and enable the synaptic plasticity that underlies learning. A diverse array of AMPAR signalling complexes are ...AMPA receptors (AMPARs) mediate the majority of excitatory transmission in the brain and enable the synaptic plasticity that underlies learning. A diverse array of AMPAR signalling complexes are established by receptor auxiliary subunits, which associate with the AMPAR in various combinations to modulate trafficking, gating and synaptic strength. However, their mechanisms of action are poorly understood. Here we determine cryo-electron microscopy structures of the heteromeric GluA1-GluA2 receptor assembled with both TARP-γ8 and CNIH2, the predominant AMPAR complex in the forebrain, in both resting and active states. Two TARP-γ8 and two CNIH2 subunits insert at distinct sites beneath the ligand-binding domains of the receptor, with site-specific lipids shaping each interaction and affecting the gating regulation of the AMPARs. Activation of the receptor leads to asymmetry between GluA1 and GluA2 along the ion conduction path and an outward expansion of the channel triggers counter-rotations of both auxiliary subunit pairs, promoting the active-state conformation. In addition, both TARP-γ8 and CNIH2 pivot towards the pore exit upon activation, extending their reach for cytoplasmic receptor elements. CNIH2 achieves this through its uniquely extended M2 helix, which has transformed this endoplasmic reticulum-export factor into a powerful AMPAR modulator that is capable of providing hippocampal pyramidal neurons with their integrative synaptic properties. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12805.map.gz emd_12805.map.gz | 7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12805-v30.xml emd-12805-v30.xml emd-12805.xml emd-12805.xml | 28 KB 28 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12805_fsc.xml emd_12805_fsc.xml emd_12805_fsc_2.xml emd_12805_fsc_2.xml emd_12805_fsc_3.xml emd_12805_fsc_3.xml | 11.3 KB 11.4 KB 11.3 KB | Display Display Display |  FSC data file FSC data file |
| Images |  emd_12805.png emd_12805.png | 202.9 KB | ||
| Filedesc metadata |  emd-12805.cif.gz emd-12805.cif.gz | 7.7 KB | ||
| Others |  emd_12805_additional_1.map.gz emd_12805_additional_1.map.gz emd_12805_additional_2.map.gz emd_12805_additional_2.map.gz emd_12805_additional_3.map.gz emd_12805_additional_3.map.gz emd_12805_additional_4.map.gz emd_12805_additional_4.map.gz | 11 MB 1.4 MB 8.8 MB 7.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12805 http://ftp.pdbj.org/pub/emdb/structures/EMD-12805 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12805 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12805 | HTTPS FTP |
-Related structure data
| Related structure data |  7oceMC  7ocaC  7occC  7ocdC  7ocfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12805.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12805.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map generated based on masked refinement maps of LBD and TMD. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Masked refinement map focusing on TMD
| File | emd_12805_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked refinement map focusing on TMD | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Masked refinement map focusing on CNIH2
| File | emd_12805_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked refinement map focusing on CNIH2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Masked refinement map focusing on LBD
| File | emd_12805_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked refinement map focusing on LBD | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: EMDA best map generated based on masked refinement...
| File | emd_12805_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EMDA best map generated based on masked refinement maps of LBD and TMD. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GluA1/A2 heterotetramer in complex with auxiliary subunits TARP g...
| Entire | Name: GluA1/A2 heterotetramer in complex with auxiliary subunits TARP gamma 8 and CNIH2 |
|---|---|
| Components |
|
-Supramolecule #1: GluA1/A2 heterotetramer in complex with auxiliary subunits TARP g...
| Supramolecule | Name: GluA1/A2 heterotetramer in complex with auxiliary subunits TARP gamma 8 and CNIH2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 527 KDa |
-Macromolecule #1: Glutamate receptor 1
| Macromolecule | Name: Glutamate receptor 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 102.66193 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPYIFAFFCT GFLGAVVGAD YKDDDDKNFP NNIQIGGLFP NQQSQEHAAF RFALSQLTEP PKLLPQIDIV NISDSFEMTY RFCSQFSKG VYAIFGFYER RTVNMLTSFC GALHVCFITP SFPVDTSNQF VLQLRPELQE ALISIIDHYK WQTFVYIYDA D RGLSVLQR ...String: MPYIFAFFCT GFLGAVVGAD YKDDDDKNFP NNIQIGGLFP NQQSQEHAAF RFALSQLTEP PKLLPQIDIV NISDSFEMTY RFCSQFSKG VYAIFGFYER RTVNMLTSFC GALHVCFITP SFPVDTSNQF VLQLRPELQE ALISIIDHYK WQTFVYIYDA D RGLSVLQR VLDTAAEKNW QVTAVNILTT TEEGYRMLFQ DLEKKKERLV VVDCESERLN AILGQIVKLE KNGIGYHYIL AN LGFMDID LNKFKESGAN VTGFQLVNYT DTIPARIMQQ WRTSDSRDHT RVDWKRPKYT SALTYDGVKV MAEAFQSLRR QRI DISRRG NAGDCLANPA VPWGQGIDIQ RALQQVRFEG LTGNVQFNEK GRRTNYTLHV IEMKHDGIRK IGYWNEDDKF VPAA TDAQA GGDNSSVQNR TYIVTTILED PYVMLKKNAN QFEGNDRYEG YCVELAAEIA KHVGYSYRLE IVSDGKYGAR DPDTK AWNG MVGELVYGRA DVAVAPLTIT LVREEVIDFS KPFMSLGISI MIKKPQKSKP GVFSFLDPLA YEIWMCIVFA YIGVSV VLF LVSRFSPYEW HSEEFEEGRD QTTSDQSNEF GIFNSLWFSL GAFMQQGCDI SPRSLSGRIV GGVWWFFTLI IISSYTA NL AAFLTVERMV SPIESAEDLA KQTEIAYGTL EAGSTKEFFR RSKIAVFEKM WTYMKSAEPS VFVRTTEEGM IRVRKSKG K YAYLLESTMN EYIEQRKPCD TMKVGGNLDS KGYGIATPKG SALRGPVNLA VLKLSEQGVL DKLKSKWWYD KGECGSKDS GSKDKTSALS LSNVAGVFYI LIGGLGLAML VALIEFCYKS RSESKRMKGF CLIPQQSINE AIRTSTLPRN SGAGASGGGG SGENGRVVS QDFPKSMQSI PCMSHSSGMP LGATGL UniProtKB: Glutamate receptor 1 |
-Macromolecule #2: Glutamate receptor 2
| Macromolecule | Name: Glutamate receptor 2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 96.247055 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MQKIMHISVL LSPVLWGLIF GVSSNSIQIG GLFPRGADQE YSAFRVGMVQ FSTSEFRLTP HIDNLEVANS FAVTNAFCSQ FSRGVYAIF GFYDKKSVNT ITSFCGTLHV SFITPSFPTD GTHPFVIQMR PDLKGALLSL IEYYQWDKFA YLYDSDRGLS T LQAVLDSA ...String: MQKIMHISVL LSPVLWGLIF GVSSNSIQIG GLFPRGADQE YSAFRVGMVQ FSTSEFRLTP HIDNLEVANS FAVTNAFCSQ FSRGVYAIF GFYDKKSVNT ITSFCGTLHV SFITPSFPTD GTHPFVIQMR PDLKGALLSL IEYYQWDKFA YLYDSDRGLS T LQAVLDSA AEKKWQVTAI NVGNINNDKK DETYRSLFQD LELKKERRVI LDCERDKVND IVDQVITIGK HVKGYHYIIA NL GFTDGDL LKIQFGGANV SGFQIVDYDD SLVSKFIERW STLEEKEYPG AHTATIKYTS ALTYDAVQVM TEAFRNLRKQ RIE ISRRGN AGDCLANPAV PWGQGVEIER ALKQVQVEGL SGNIKFDQNG KRINYTINIM ELKTNGPRKI GYWSEVDKMV VTLT ELPSG NDTSGLENKT VVVTTILESP YVMMKKNHEM LEGNERYEGY CVDLAAEIAK HCGFKYKLTI VGDGKYGARD ADTKI WNGM VGELVYGKAD IAIAPLTITL VREEVIDFSK PFMSLGISIM IKKPQKSKPG VFSFLDPLAY EIWMCIVFAY IGVSVV LFL VSRFSPYEWH TEEFEDGRET QSSESTNEFG IFNSLWFSLG AFMRQGCDIS PRSLSGRIVG GVWWFFTLII ISSYTAN LA AFLTVERMVS PIESAEDLSK QTEIAYGTLD SGSTKEFFRR SKIAVFDKMW TYMRSAEPSV FVRTTAEGVA RVRKSKGK Y AYLLESTMNE YIEQRKPCDT MKVGGNLDSK GYGIATPKGS SLGTPVNLAV LKLSEQGVLD KLKNKWWYDK GECGAKDSG SKEKTSALSL SNVAGVFYIL VGGLGLAMLV ALIEFCYKSR AEAKRMKVAK NPQNINPSSS UniProtKB: Glutamate receptor 2 |
-Macromolecule #3: Protein cornichon homolog 2
| Macromolecule | Name: Protein cornichon homolog 2 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.000605 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAFTFAAFCY MLTLVLCASL IFFVIWHIIA FDELRTDFKN PIDQGNPARA RERLKNIERI CCLLRKLVVP EYSIHGLFCL MFLCAAEWV TLGLNIPLLF YHLWRYFHRP ADGSEVMYDA VSIMNADILN YCQKESWCKL AFYLLSFFYY LYSMVYTLVS F ENLYFQSG GSTETSQVAP AYPYDVPDYA UniProtKB: Protein cornichon homolog 2 |
-Macromolecule #4: Voltage-dependent calcium channel gamma-8 subunit
| Macromolecule | Name: Voltage-dependent calcium channel gamma-8 subunit / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.518957 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ESLKRWNEER GLWCEKGVQV LLTTIGAFAA FGLMTIAIST DYWLYTRALI CNTTNLTAGD DGPPHRGGSG SSEKKDPGGL THSGLWRIC CLEGLKRGVC VKINHFPEDT DYDHDSAEYL LRVVRASSIF PILSAILLLL GGVCVAASRV YKSKRNIILG A GILFVAAG ...String: ESLKRWNEER GLWCEKGVQV LLTTIGAFAA FGLMTIAIST DYWLYTRALI CNTTNLTAGD DGPPHRGGSG SSEKKDPGGL THSGLWRIC CLEGLKRGVC VKINHFPEDT DYDHDSAEYL LRVVRASSIF PILSAILLLL GGVCVAASRV YKSKRNIILG A GILFVAAG LSNIIGVIVY ISANAGEPGP KRDEEKKNHY SYGWSFYFGG LSFILAEVIG VLAVNIYIER SREAHCQSRS DL LKAGGGA GGSGGSGPSA ILRLPSYRFR YRRRSRSSSR GSSEASPSRD ASPGGPGGPG FASTDISMYT LSRDPSKGSV AAG LASAGG GGGGAGVGAY GGAAGAAGGG GTGSERDRGS SAGFLTLHNA FPKEAASGVT VTVTGPPAAP APAPPAPAAP APGT LSKEA AASNTNTLNR KLEVLFQ UniProtKB: Voltage-dependent calcium channel gamma-8 subunit |
-Macromolecule #5: 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-s...
| Macromolecule | Name: 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide type: ligand / ID: 5 / Number of copies: 4 / Formula: E2Q |
|---|---|
| Molecular weight | Theoretical: 336.28 Da |
| Chemical component information | 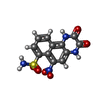 ChemComp-E2Q: |
-Macromolecule #6: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 6 / Number of copies: 46 / Formula: PC1 |
|---|---|
| Molecular weight | Theoretical: 790.145 Da |
| Chemical component information |  ChemComp-PC1: |
-Macromolecule #7: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 7 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | Purified protein was incubated with 100 uM NBQX for at least 30 min on ice before freezing |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 2.5 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 113 | ||||||
| Output model |  PDB-7oce: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)