+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12018 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | VAR2CSA full ectodomain in present of plCS, DBL1-DBL4 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | VAR2CSA / CELL ADHESION / malaria / pfEMP1 / DBL | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
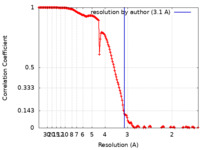
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Wang KT / Dagil R | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Cryo-EM reveals the architecture of placental malaria VAR2CSA and provides molecular insight into chondroitin sulfate binding. Authors: Kaituo Wang / Robert Dagil / Thomas Lavstsen / Sandeep K Misra / Charlotte B Spliid / Yong Wang / Tobias Gustavsson / Daniel R Sandoval / Elena Ethel Vidal-Calvo / Swati Choudhary / Mette Ø ...Authors: Kaituo Wang / Robert Dagil / Thomas Lavstsen / Sandeep K Misra / Charlotte B Spliid / Yong Wang / Tobias Gustavsson / Daniel R Sandoval / Elena Ethel Vidal-Calvo / Swati Choudhary / Mette Ø Agerbaek / Kresten Lindorff-Larsen / Morten A Nielsen / Thor G Theander / Joshua S Sharp / Thomas Mandel Clausen / Pontus Gourdon / Ali Salanti /    Abstract: Placental malaria can have severe consequences for both mother and child and effective vaccines are lacking. Parasite-infected red blood cells sequester in the placenta through interaction between ...Placental malaria can have severe consequences for both mother and child and effective vaccines are lacking. Parasite-infected red blood cells sequester in the placenta through interaction between parasite-expressed protein VAR2CSA and the glycosaminoglycan chondroitin sulfate A (CS) abundantly present in the intervillous space. Here, we report cryo-EM structures of the VAR2CSA ectodomain at up to 3.1 Å resolution revealing an overall V-shaped architecture and a complex domain organization. Notably, the surface displays a single significantly electropositive patch, compatible with binding of negatively charged CS. Using molecular docking and molecular dynamics simulations as well as comparative hydroxyl radical protein foot-printing of VAR2CSA in complex with placental CS, we identify the CS-binding groove, intersecting with the positively charged patch of the central VAR2CSA structure. We identify distinctive conserved structural features upholding the macro-molecular domain complex and CS binding capacity of VAR2CSA as well as divergent elements possibly allowing immune escape at or near the CS binding site. These observations will support rational design of second-generation placental malaria vaccines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12018.map.gz emd_12018.map.gz | 162.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12018-v30.xml emd-12018-v30.xml emd-12018.xml emd-12018.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12018_fsc.xml emd_12018_fsc.xml | 15.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_12018.png emd_12018.png | 61.6 KB | ||
| Filedesc metadata |  emd-12018.cif.gz emd-12018.cif.gz | 6.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12018 http://ftp.pdbj.org/pub/emdb/structures/EMD-12018 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12018 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12018 | HTTPS FTP |
-Related structure data
| Related structure data |  7b54MC  7b52C  7nnhC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12018.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12018.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : VAR2CSA in presence of plCS, DBl1-DBL4
| Entire | Name: VAR2CSA in presence of plCS, DBl1-DBL4 |
|---|---|
| Components |
|
-Supramolecule #1: VAR2CSA in presence of plCS, DBl1-DBL4
| Supramolecule | Name: VAR2CSA in presence of plCS, DBl1-DBL4 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: VAR2CSA in presence of plCS, DBl1-DBL4,Erythrocyte membrane protein 1
| Macromolecule | Name: VAR2CSA in presence of plCS, DBl1-DBL4,Erythrocyte membrane protein 1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 219.438562 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MDSTSTIANK IEEYLGAKSD DSKIDELLKA DPSEVEYYRS GGDGDYLKNN ICKITVNHSD SGKYDPCEKK LPPYDDNDQW KCQQNSSDG SGKPENICVP PRRERLCTYN LENLKFDKIR DNNAFLADVL LTARNEGEKI VQNHPDTNSS NVCNALERSF A DLADIIRG ...String: MDSTSTIANK IEEYLGAKSD DSKIDELLKA DPSEVEYYRS GGDGDYLKNN ICKITVNHSD SGKYDPCEKK LPPYDDNDQW KCQQNSSDG SGKPENICVP PRRERLCTYN LENLKFDKIR DNNAFLADVL LTARNEGEKI VQNHPDTNSS NVCNALERSF A DLADIIRG TDQWKGTNSN LEKNLKQMFA KIRENDKVLQ DKYPKDQKYT KLREAWWNAN RQKVWEVITC GARSNDLLIK RG WRTSGKS DRKKNFELCR KCGHYEKEVP TKLDYVPQFL RWLTEWIEDF YREKQNLIDD MERHREECTR EDHKSKEGTS YCS TCKDKC KKYCECVKKW KTEWENQENK YKDLYEQNKN KTSQKNTSRY DDYVKDFFEK LEANYSSLEN YIKGDPYFAE YATK LSFIL NPSDGILQEN CSDNKRGSSS NDSCDNKNQD ECQKKLEKVF ASLTNGYKCD KCKSGTSRSK KKWIWKKSSG NEEGL QEEY ANTIGLPPRT QSLYLGNLPK LENVCEDVKD INFDTKEKFL AGCLIVSFHE GKNLKKRYPQ NKNSGNKENL CKALEY SFA DYGDLIKGTS IWDNEYTKDL ELNLQNNFGK LFGKYIKKNN TAEQDTSYSS LDELRESWWN TNKKYIWTAM KHGAEMN IT TCNADGSVTG SGSSCDDIPT IDLIPQYLRF LQEWVENFCE QRQAKVKDVI TNCKSCKESG NKCKTECKTK CKDECEKY K KFIEACGTAG GGIGTAGSPW SKRWDQIYKR YSKHIEDAKR NRKAGTKNCG TSSTTNAAAS TDENKCVQSD IDSFFKHLI DIGLTTPSSY LSNVLDDNIC GADKAPWTTY TTYTTTEKCN KERDKSKSQS SDTLVVVNVP SPLGNTPYRY KYACQCKIPT NEETCDDRK EYMNQWSCGS ARTMKRGYKN DNYELCKYNG VDVKPTTVRS NSSKLDGNDV TFFNLFEQWN KEIQYQIEQY M TNANISCI DEKEVLDSVS DEGTPKVRGG YEDGRNNNTD QGTNCKEKCK CYKLWIEKIN DQWGKQKDNY NKFRSKQIYD AN KGSQNKK VVSLSNFLFF SCWEEYIQKY FNGDWSKIKN IGSDTFEFLI KKCGNNSAHG EEIFSEKLKN AEKKCKENES TDT NINKSE TSCDLNATNY IRGCQSKTYD GKIFPGKGGE KQWICKDTII HGDTNGACIP PRTQNLCVGE LWDKSYGGRS NIKN DTKEL LKEKIKNAIH KETELLYEYH DTGTAIISKN DKKGQKGKND PNGLPKGFCH AVQRSFIDYK NMILGTSVNI YEHIG KLQE DIKKIIEKGT PQQKDKIGGV GSSTENVNAW WKGIEREMWD AVRCAITKIN KKNNNSIFNG DECGVSPPTG NDEDQS VSW FKEWGEQFCI ERLRYEQNIR EACTINGKNE KKCINSKSGQ GDKIQGACKR KCEKYKKYIS EKKQEWDKQK TKYENKY VG KSASDLLKEN YPECISANFD FIFNDNIEYK TYYPYGDYSS ICSCEQVKYY KYNNAEKKNN KSLCYEKDND MTWSKKYI K KLENGRSLEG VYVPPRRQQL CLYELFPIII KNEEGMEKAK EELLETLQIV AEREAYYLWK QYNPTGKGID DANKKACCA IRGSFYDLED IIKGNDLVHD EYTKYIDSKL NEIFGSSNTN DIDTKRARTD WWENETITNG TDRKTIRQLV WDAMQSGVRY AVEEKNENF PLCMGVEHIG IAKPQFIRWL EEWTNEFCEK YTKYFEDMKS KCDPPKRADT CGDNSNIECK KACANYTNWL N PKRIEWNG MSNYYNKIYR KSNKESEDGK DYSMIMAPTV IDYLNKRCHG EINGNYICCS CKNIGAYNTT SGTVNKKLQK KE TECEEEK GPLDLMNEVL NKMDKKYSAH KMKCTEVYLE HVEEQLNEID NAIKDYKL UniProtKB: Erythrocyte membrane protein 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20mM Tris pH 7.5 and 75mM KCl |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7b54: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)