+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11777 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | RET/GDF15/GFRAL extracellular complex negative stain envelope | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 26.0 Å | |||||||||
 Authors Authors | Adams SE / Earl CP / McDonald NQ | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2021 Journal: Structure / Year: 2021Title: A two-site flexible clamp mechanism for RET-GDNF-GFRα1 assembly reveals both conformational adaptation and strict geometric spacing. Authors: Sarah E Adams / Andrew G Purkiss / Phillip P Knowles / Andrea Nans / David C Briggs / Annabel Borg / Christopher P Earl / Kerry M Goodman / Agata Nawrotek / Aaron J Borg / Pauline B McIntosh ...Authors: Sarah E Adams / Andrew G Purkiss / Phillip P Knowles / Andrea Nans / David C Briggs / Annabel Borg / Christopher P Earl / Kerry M Goodman / Agata Nawrotek / Aaron J Borg / Pauline B McIntosh / Francesca M Houghton / Svend Kjær / Neil Q McDonald /  Abstract: RET receptor tyrosine kinase plays vital developmental and neuroprotective roles in metazoans. GDNF family ligands (GFLs) when bound to cognate GFRα co-receptors recognize and activate RET ...RET receptor tyrosine kinase plays vital developmental and neuroprotective roles in metazoans. GDNF family ligands (GFLs) when bound to cognate GFRα co-receptors recognize and activate RET stimulating its cytoplasmic kinase function. The principles for RET ligand-co-receptor recognition are incompletely understood. Here, we report a crystal structure of the cadherin-like module (CLD1-4) from zebrafish RET revealing interdomain flexibility between CLD2 and CLD3. Comparison with a cryo-electron microscopy structure of a ligand-engaged zebrafish RET-GDNF-GFRα1a complex indicates conformational changes within a clade-specific CLD3 loop adjacent to the co-receptor. Our observations indicate that RET is a molecular clamp with a flexible calcium-dependent arm that adapts to different GFRα co-receptors, while its rigid arm recognizes a GFL dimer to align both membrane-proximal cysteine-rich domains. We also visualize linear arrays of RET-GDNF-GFRα1a suggesting that a conserved contact stabilizes higher-order species. Our study reveals that ligand-co-receptor recognition by RET involves both receptor plasticity and strict spacing of receptor dimers by GFL ligands. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11777.map.gz emd_11777.map.gz | 4.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11777-v30.xml emd-11777-v30.xml emd-11777.xml emd-11777.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11777.png emd_11777.png | 65.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11777 http://ftp.pdbj.org/pub/emdb/structures/EMD-11777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11777 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11777.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11777.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.438 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
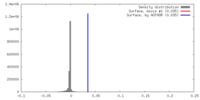
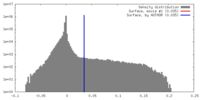
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ternary complex of the human RET extracellular domain with the GD...
| Entire | Name: Ternary complex of the human RET extracellular domain with the GDF15/GFRAL ligand-co-receptor pair |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of the human RET extracellular domain with the GD...
| Supramolecule | Name: Ternary complex of the human RET extracellular domain with the GDF15/GFRAL ligand-co-receptor pair type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: Freestyle 293-F / Recombinant plasmid: pCEP Homo sapiens (human) / Recombinant cell: Freestyle 293-F / Recombinant plasmid: pCEP |
| Molecular weight | Theoretical: 240 KDa |
-Macromolecule #1: Proto-oncogene tyrosine-protein kinase receptor Ret
| Macromolecule | Name: Proto-oncogene tyrosine-protein kinase receptor Ret / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: LYFSRDAYWE KLYVDQAAGT PLLYVHALRD APEEVPSFRL GQHLYGTYRT RLHENNWIRI QEDTGLLYLQ RSLDHSSWEK LSVRNRGFPL LTVYLKVFLS PTSLREGECQ WPGCARVYFS FFNTSFPACS SLKPRELCFP ETRPSFRIRE NRPPGTFHQF RLLPVQFLCP ...String: LYFSRDAYWE KLYVDQAAGT PLLYVHALRD APEEVPSFRL GQHLYGTYRT RLHENNWIRI QEDTGLLYLQ RSLDHSSWEK LSVRNRGFPL LTVYLKVFLS PTSLREGECQ WPGCARVYFS FFNTSFPACS SLKPRELCFP ETRPSFRIRE NRPPGTFHQF RLLPVQFLCP QISVAYRLLE GEGLPFRSAP DSLEVSTRWA LDREQREKYE LVAVCTVHAG AREEVVMVPF PVTVYDEDDS APTFPAGVDT ASAVVEFKRK EDTVVATLRV FDADVVPASG ELVRRYTSTL LPGDTWAQQT FRVEHWPNET SVQANGSFVR ATVHDYRLVL NRNLSISENR TMQLAVLVND SDFQGPGAGV LLLHFNVSVL PVSLHLPSTY SLSVSRRARR FAQIGKVCVE NCQAFSGINV QYKLHSSGAN CSTLGVVTSA EDTSGILFVN DTKALRRPKC AELHYMVVAT DQQTSRQAQA QLLVTVEGSY VAEEAGCPLS CAVSKRRLEC EECGGLGSPT GRCEWRQGDG KGITRNFSTC SPSTKTCPDG HCDVVETQDI NICPQDCLRG SIVGGHEPGE PRGIKAGYGT CNCFPEEEKC FCEPEDIQDP LCDELCRTGE NLYF |
-Macromolecule #2: GDNF family receptor alpha-like
| Macromolecule | Name: GDNF family receptor alpha-like / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: APLANNCTYL REQCLRDANG CKHAWRVMED ACNDSDPGDP CKMRNSSYCN LSIQYLVESN FQFKECLCTD DFYCTVNKLL GKKCINKSDN VKEDKFKWNL TTRSHHGFKG MWSCLEVAEA CVGDVVCNAQ LASYLKACSA NGNPCDLKQC QAAIRFFYQN IPFNIAQMLA ...String: APLANNCTYL REQCLRDANG CKHAWRVMED ACNDSDPGDP CKMRNSSYCN LSIQYLVESN FQFKECLCTD DFYCTVNKLL GKKCINKSDN VKEDKFKWNL TTRSHHGFKG MWSCLEVAEA CVGDVVCNAQ LASYLKACSA NGNPCDLKQC QAAIRFFYQN IPFNIAQMLA FCDCAQSDIP CQQSKEALHS KTCAVNMVPP PTCLSVIRSC QNDELCRRHY RTFQSKCWQR VTRKCHEDEN CISTLSKQDL TCSGSDDCKA AYIDILGTVL QVQCTCRTIT QSEESLCKIF QHMLHRKSCF NYPTLSNVKG MALYTRKHAN KITLTGFHSP FNGEAAAHHH HHH |
-Macromolecule #3: Growth/differentiation factor 15
| Macromolecule | Name: Growth/differentiation factor 15 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: APLARARNGD HCPLGPGRCC RLHTVRASLE DLGWADWVLS PREVQVTMCI GACPS QFRA ANMHAQIKTS LHRLKPDTVP APCCVPASYN PMVLIQKTDT GVSLQTYDDL LAKDCH CI |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 Component:
Details: Protein crosslinked by GraFIX method with sucrose/gluteraldehyde gradient | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Staining | Type: NEGATIVE / Material: uranyl acetate | ||||||||||||||||||
| Grid | Model: Homemade / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Details: 45 mA |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 1000 (2k x 2k) / Number grids imaged: 1 / Number real images: 299 / Average exposure time: 1.0 sec. / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 42000 |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)