+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11712 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
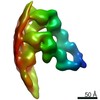
| Title | HDAC-DC-nucleosome | |||||||||
 Map data Map data | HDAC-DC-nucleosome | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 10.6 Å | |||||||||
 Authors Authors | Lee J-H / Bollschweiler D / Schaefer T / Huber R | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies. Authors: Jung-Hoon Lee / Daniel Bollschweiler / Tillman Schäfer / Robert Huber /  Abstract: The chromatin-modifying histone deacetylases (HDACs) remove acetyl groups from acetyl-lysine residues in histone amino-terminal tails, thereby mediating transcriptional repression. Structural makeup ...The chromatin-modifying histone deacetylases (HDACs) remove acetyl groups from acetyl-lysine residues in histone amino-terminal tails, thereby mediating transcriptional repression. Structural makeup and mechanisms by which multisubunit HDAC complexes recognize nucleosomes remain elusive. Our cryo-electron microscopy structures of the yeast class II HDAC ensembles show that the HDAC protomer comprises a triangle-shaped assembly of stoichiometry Hda1-Hda2-Hda3, in which the active sites of the Hda1 dimer are freely accessible. We also observe a tetramer of protomers, where the nucleosome binding modules are inaccessible. Structural analysis of the nucleosome-bound complexes indicates how positioning of Hda1 adjacent to histone H2B affords HDAC catalysis. Moreover, it reveals how an intricate network of multiple contacts between a dimer of protomers and the nucleosome creates a platform for expansion of the HDAC activities. Our study provides comprehensive insight into the structural plasticity of the HDAC complex and its functional mechanism of chromatin modification. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11712.map.gz emd_11712.map.gz | 31.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11712-v30.xml emd-11712-v30.xml emd-11712.xml emd-11712.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11712.png emd_11712.png | 49.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11712 http://ftp.pdbj.org/pub/emdb/structures/EMD-11712 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11712 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11712 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11712.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11712.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HDAC-DC-nucleosome | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.885 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HDAC-PC-Nuc
| Entire | Name: HDAC-PC-Nuc |
|---|---|
| Components |
|
-Supramolecule #1: HDAC-PC-Nuc
| Supramolecule | Name: HDAC-PC-Nuc / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#14 |
|---|
-Supramolecule #2: Histone deacetylase
| Supramolecule | Name: Histone deacetylase / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Supramolecule #3: Histone
| Supramolecule | Name: Histone / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #5-#12 |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  |
-Supramolecule #4: DNA
| Supramolecule | Name: DNA / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #13-#14 |
|---|---|
| Source (natural) | Organism: unidentified plasmid (others) |
| Recombinant expression | Organism: synthetic construct (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.8 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 10.6 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cisTEM (ver. 1) / Number images used: 9534 |
|---|---|
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
-Atomic model buiding 1
| Details | Real space refinement |
|---|
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















