Yorodumi
Yorodumi+ Open data
Open data
 Loading...
Loading...
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10985 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of the mitoribosome from Neurospora crassa with bound tRNA at the P-site | |||||||||||||||
 Map data Map data | Composite map for the P-tRNA bound state. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Neurospora crassa / translating Mitoribosomes / tRNA / mRNA / mL108 / TRANSLATION | |||||||||||||||
| Function / homology |  Function and homology information Function and homology information3-hydroxyisobutyryl-CoA hydrolase / 3-hydroxyisobutyryl-CoA hydrolase activity / L-valine catabolic process / : / negative regulation of ATP-dependent activity / ribonuclease III activity / ATPase inhibitor activity / DNA strand exchange activity / mitochondrial large ribosomal subunit / mitochondrial ribosome ...3-hydroxyisobutyryl-CoA hydrolase / 3-hydroxyisobutyryl-CoA hydrolase activity / L-valine catabolic process / : / negative regulation of ATP-dependent activity / ribonuclease III activity / ATPase inhibitor activity / DNA strand exchange activity / mitochondrial large ribosomal subunit / mitochondrial ribosome / mitochondrial small ribosomal subunit / mitochondrial translation / superoxide dismutase activity / positive regulation of proteolysis / RNA processing / rRNA processing / peroxisome / single-stranded DNA binding / double-stranded RNA binding / large ribosomal subunit / transferase activity / small ribosomal subunit / small ribosomal subunit rRNA binding / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mitochondrion / RNA binding / metal ion binding / cytoplasm Similarity search - Function 54S ribosomal protein L44, mitochondrial / : / Ribosomal protein L31, mitochondrial / Ribosomal protein L20, mitochondrial / Mitochondrial homologous recombination protein 1 / 54S ribosomal protein L25 / 54S ribosomal protein L15, mitochondrial / MRPL25 domain / 54S ribosomal protein L3, double-stranded RNA binding domain / Mitochondrial ribosomal protein subunit L20 ...54S ribosomal protein L44, mitochondrial / : / Ribosomal protein L31, mitochondrial / Ribosomal protein L20, mitochondrial / Mitochondrial homologous recombination protein 1 / 54S ribosomal protein L25 / 54S ribosomal protein L15, mitochondrial / MRPL25 domain / 54S ribosomal protein L3, double-stranded RNA binding domain / Mitochondrial ribosomal protein subunit L20 / Transcriptional regulation of mitochondrial recombination / Mitochondrial ribosomal protein mL59 / Mitochondrial ribosomal protein L31 / 54S ribosomal protein L36, yeast / 54S ribosomal protein L28, mitochondrial / : / Ribosomal protein L31p, N-terminal / Mitochondrial ribosomal protein MRP51, fungi / Mitochondrial ribosomal protein S25 / Ribosomal protein S24, mitochondrial / Ribosomal protein S23, mitochondrial, fungi / Mitochondrial ribosomal protein subunit / Mitochondrial ribosomal protein S25 / Ribosomal protein S35, mitochondrial / Enoyl-CoA hydratase/isomerase, HIBYL-CoA-H type / Ribosomal protein MRP10, mitochondrial / Eukaryotic mitochondrial regulator protein / Enoyl-CoA hydratase/isomerase domain / Enoyl-CoA hydratase/isomerase / Protein Fyv4 / Small ribosomal subunit protein mS41 SAM domain / IGR protein motif / IGR / Ribonuclease III / Ribonuclease-III-like / Mitochondrial ATPase inhibitor / Mitochondrial ATPase inhibitor, IATP / 28S ribosomal protein S25, mitochondrial / Ribonuclease III family domain profile. / Ribonuclease III family / Ribonuclease III domain / Ribosomal protein L53, mitochondrial / 39S ribosomal protein L53/MRP-L53 / Ribosomal protein S27/S33, mitochondrial / Ribosomal protein S24/S35, mitochondrial / Mitochondrial ribosomal subunit S27 / Ribosomal protein L37, mitochondrial / Mitochondrial ribosomal protein L37 / Ribosomal protein S24/S35, mitochondrial, conserved domain / Mitochondrial ribosomal subunit protein / Enoyl-CoA hydratase/isomerase, conserved site / Enoyl-CoA hydratase/isomerase signature. / Ribosomal protein S23/S29, mitochondrial / Mitochondrial ribosomal death-associated protein 3 / Ribosomal protein L50, mitochondria / Ribosomal subunit 39S / Ribosomal protein L46, N-terminal / 39S mitochondrial ribosomal protein L46 / Ribosomal protein L49/IMG2 / Mitochondrial large subunit ribosomal protein (Img2) / Ribosomal protein L27/L41, mitochondrial / : / Mitochondrial ribosomal protein L27 / 39S ribosomal protein L46, mitochondrial / Mitochondrial mRNA-processing protein COX24, C-terminal / Mitochondrial mRNA-processing protein COX24, C-terminal / Mitochondrial domain of unknown function (DUF1713) / Ribosomal protein L47, mitochondrial / Manganese/iron superoxide dismutase, C-terminal / Manganese/iron superoxide dismutase, C-terminal domain superfamily / Manganese/iron superoxide dismutase, N-terminal domain superfamily / MRP-L47 superfamily, mitochondrial / 39S ribosomal protein L43/54S ribosomal protein L51 / Iron/manganese superoxide dismutases, C-terminal domain / Mitochondrial 39-S ribosomal protein L47 (MRP-L47) / Phosphatidylethanolamine-binding protein, eukaryotic / Phosphatidylethanolamine-binding protein / Phosphatidylethanolamine-binding protein / PEBP-like superfamily / Double-stranded RNA binding motif / Ribonuclease III, endonuclease domain superfamily / Double-stranded RNA-binding domain / : / Mitochondrial ribosomal protein L51 / S25 / CI-B8 domain / Ribosomal protein/NADH dehydrogenase domain / Mitochondrial ribosomal protein L51 / S25 / CI-B8 domain / ClpP/crotonase-like domain superfamily / Ribosomal protein L1, 3-layer alpha/beta-sandwich / NUDIX hydrolase-like domain superfamily / Ribosomal protein L1-like / Ribosomal protein L1/ribosomal biogenesis protein / Ribosomal protein L1p/L10e family / Ribosomal protein L11, bacterial-type / Sterile alpha motif/pointed domain superfamily / Ribosomal protein S21 / Ribosomal protein S16, conserved site / Ribosomal protein S16 signature. / Ribosomal protein S21 / Ribosomal protein L10-like domain superfamily / Ribosomal protein L16 signature 2. Similarity search - Domain/homology Mitochondrial 37S ribosomal protein MRP2 / ATP synthase F1 subunit epsilon / Nucleic acid-binding protein / 60S ribosomal protein L2, mitochondrial / 37S ribosomal protein mrp5 / RIBOSOMAL_L9 domain-containing protein / Ribosomal protein S16 / Ribosomal protein L15 / Manganese and iron superoxide dismutase / Large ribosomal subunit protein mL59 domain-containing protein ...Mitochondrial 37S ribosomal protein MRP2 / ATP synthase F1 subunit epsilon / Nucleic acid-binding protein / 60S ribosomal protein L2, mitochondrial / 37S ribosomal protein mrp5 / RIBOSOMAL_L9 domain-containing protein / Ribosomal protein S16 / Ribosomal protein L15 / Manganese and iron superoxide dismutase / Large ribosomal subunit protein mL59 domain-containing protein / Mitochondrial 54S ribosomal protein YmL19 / 54S ribosomal protein L24 / Ribosomal protein S6 / Ribonuclease III / KOW domain-containing protein / Ribosomal protein S12 / Translational machinery component / Ribosomal_L6 domain-containing protein / Ribosomal protein S19/S15 / Mitochondrial ribosomal protein subunit L23 / Ribosomal protein S21 / MRP-L46 domain-containing protein / Mitochondrial 37S ribosomal protein S27 / L51_S25_CI-B8 domain-containing protein / 54S ribosomal protein L4, mitochondrial / Uncharacterized protein / 54S ribosomal protein IMG1, mitochondrial / S4 domain-containing protein / Protein FYV4, mitochondrial / PEBP-like protein / Mitochondrial ribosomal protein L37-domain-containing protein / 37S ribosomal protein mrp10, mitochondrial / Ribosomal protein L13 / 54S ribosomal protein L27, mitochondrial / RNase III domain-containing protein / Ribosomal protein L33 / Ribosomal protein L14 / Ribosomal_S7 domain-containing protein / 30S ribosomal protein S10, mitochondrial / Mitochondrial large ribosomal subunit protein / Ribosomal protein L17 / 37S ribosomal protein S9, mitochondrial / 37S ribosomal protein S8, mitochondrial / 54S ribosomal protein L31, mitochondrial / Ribosomal protein S5 / Related to ribosomal protein YmL11, mitochondrial / Large ribosomal subunit protein bL21m / Large ribosomal subunit protein mL53 / Small ribosomal subunit protein uS13m / Large ribosomal subunit protein mL61 / Small ribosomal subunit protein mS27 / Large ribosomal subunit protein bL36m / Large ribosomal subunit protein mL49 / Large ribosomal subunit protein uL22m / Large ribosomal subunit protein mL50 / Small ribosomal subunit protein mS29 / Small ribosomal subunit protein mS45 / Small ribosomal subunit protein mS38 / Small ribosomal subunit protein mS26 / Related to ribosomal protein L1 / Small ribosomal subunit protein bS18m / Large ribosomal subunit protein uL3m / Small ribosomal subunit protein mS23 / Related to ribosomal protein YmL20, mitochondrial / Small ribosomal subunit protein uS5m / Related to ribosomal protein S15 (Mitochondrial) / Related to ribosomal protein L16, mitochondrial / Large ribosomal subunit protein uL30m / Large ribosomal subunit protein bL27m / Large ribosomal subunit protein uL5m / Related to ribosomal protein L34, mitochondrial / 3-hydroxyisobutyryl-CoA hydrolase / Related to ribosomal protein YmL36, mitochondrial / Small ribosomal subunit protein mS41 / Large ribosomal subunit protein bL32m / Small ribosomal subunit protein mS35 / Small ribosomal subunit protein uS2m / Large ribosomal subunit protein uL4m Similarity search - Component | |||||||||||||||
| Biological species |  Neurospora crassa (fungus) Neurospora crassa (fungus) | |||||||||||||||
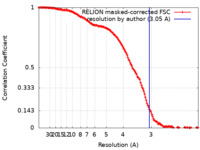
| Method | single particle reconstruction / cryo EM / Resolution: 3.05 Å | |||||||||||||||
 Authors Authors | Amunts A / Itoh Y | |||||||||||||||
| Funding support |  Sweden, European Union, 4 items Sweden, European Union, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi. Authors: Yuzuru Itoh / Andreas Naschberger / Narges Mortezaei / Johannes M Herrmann / Alexey Amunts /   Abstract: Mitoribosomes are specialized protein synthesis machineries in mitochondria. However, how mRNA binds to its dedicated channel, and tRNA moves as the mitoribosomal subunit rotate with respect to each ...Mitoribosomes are specialized protein synthesis machineries in mitochondria. However, how mRNA binds to its dedicated channel, and tRNA moves as the mitoribosomal subunit rotate with respect to each other is not understood. We report models of the translating fungal mitoribosome with mRNA, tRNA and nascent polypeptide, as well as an assembly intermediate. Nicotinamide adenine dinucleotide (NAD) is found in the central protuberance of the large subunit, and the ATPase inhibitory factor 1 (IF) in the small subunit. The models of the active mitoribosome explain how mRNA binds through a dedicated protein platform on the small subunit, tRNA is translocated with the help of the protein mL108, bridging it with L1 stalk on the large subunit, and nascent polypeptide paths through a newly shaped exit tunnel involving a series of structural rearrangements. An assembly intermediate is modeled with the maturation factor Atp25, providing insight into the biogenesis of the mitoribosomal large subunit and translation regulation. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10985.map.gz emd_10985.map.gz | 144.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10985-v30.xml emd-10985-v30.xml emd-10985.xml emd-10985.xml | 116.4 KB 116.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10985_fsc.xml emd_10985_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_10985.png emd_10985.png | 81.2 KB | ||
| Masks |  emd_10985_msk_1.map emd_10985_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10985.cif.gz emd-10985.cif.gz | 25.4 KB | ||
| Others |  emd_10985_additional_1.map.gz emd_10985_additional_1.map.gz emd_10985_half_map_1.map.gz emd_10985_half_map_1.map.gz emd_10985_half_map_2.map.gz emd_10985_half_map_2.map.gz | 143.1 MB 194.3 MB 193.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10985 http://ftp.pdbj.org/pub/emdb/structures/EMD-10985 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10985 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10985 | HTTPS FTP |
-Related structure data
| Related structure data |  6ywyMC  6yw5C  6yweC  6ywsC  6ywvC  6ywxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10985.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10985.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map for the P-tRNA bound state. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10985_msk_1.map emd_10985_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
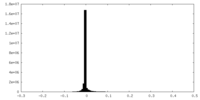
| Density Histograms |
-Additional map: Consensus map for the P-tRNA bound state.
| File | emd_10985_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus map for the P-tRNA bound state. | ||||||||||||
| Projections & Slices |
| ||||||||||||
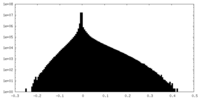
| Density Histograms |
-Half map: #2
| File | emd_10985_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
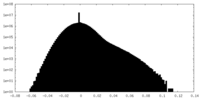
| Density Histograms |
-Half map: #1
| File | emd_10985_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Mitoribosome of Neurospora crassa in the tRNA bound state (P-site).
| Entire | Name: Mitoribosome of Neurospora crassa in the tRNA bound state (P-site). |
|---|---|
| Components |
|
+Supramolecule #1: Mitoribosome of Neurospora crassa in the tRNA bound state (P-site).
| Supramolecule | Name: Mitoribosome of Neurospora crassa in the tRNA bound state (P-site). type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#83 |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) / Strain: K5-15-23-1 Neurospora crassa (fungus) / Strain: K5-15-23-1 |
+Macromolecule #1: 23S rRNA
| Macromolecule | Name: 23S rRNA / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 1.114857375 MDa |
| Sequence | String: AAAUGUAAUG GAUAUAAAGC UUAUGUUUAU AUAUAUAGAC AUAUAUAAGU AUAUAAAGAG ACUACUACCA AUAGCUACAC UAUGUAUUA AGGAGAGUAU AACUUAAUUU AUGUUUAUGA UUUUAUCAUA CCCCUAAAAA UGACACCGAG GAGCAAGGGU C GGGUUAGC ...String: AAAUGUAAUG GAUAUAAAGC UUAUGUUUAU AUAUAUAGAC AUAUAUAAGU AUAUAAAGAG ACUACUACCA AUAGCUACAC UAUGUAUUA AGGAGAGUAU AACUUAAUUU AUGUUUAUGA UUUUAUCAUA CCCCUAAAAA UGACACCGAG GAGCAAGGGU C GGGUUAGC AUCCUGGUUC GUACACCUUG GUGACCUAGG CUAGUACCAG GUCCCCCUCU AAGGGACUUG UCCCCCUCUA AG GGACUUG CGUCGGUCCU AUCCUAGGCC GAAUAGGUGA AUAAAUACUU ACGGACGGCC UUGGUCUGUC CUAGAGGUUA UCA ACAUAU GAACUCUUAG AGAAAUUACU UAAUAAACGA AGUGAAUUGA AAUAUCUUAU UAACUUCAGG AAAAGAAAUC AAAC GAGAU UCUAUGAUUA GUGUGAACGA AAAUAGAGCA GCCUAUUAAA AUAAGUAAAA UGGCUUUAAA GCUGUUUGAA UAUUG UGGG GAACCUUCCU CAAAGGCUAA AUAUAAUACA UGAGUUACAG AGAAAAGUAC CGUGAGGGAA AGCUUUGAAA UAGUAG UUU UAUAAGCAGC UCAAGCAAUA AGAAAGCGAG AGCGUACCUU UUGCAUAAUG GGUCACCAAG UUAAUUUUAG AUGCGAG CG AAUUUAUUUA UGUUUUUACU GAUUAAACAA UAUAAUGAAU CAUAAUUAUU UUUGUAACGA GUAUUAGUAU UAAAUCUU A AUUUAAUAUU AGUAUAAGUU UUCAGUAUGG CGGCUACAUA GCAUAAUCUA UGCAGCCAGC CAAUAAUUGG AUUUCCAAU CCAAUUUCGG UAAUAAAUAG AUGUGCAUAG UUAAACCGAU CAUUAAAAUA AUGAAUAGUG UCUAAAGUUA GACCCGAAGC CUGGUGAUC UUACUAUAGU CAGGACUAUA AAGGUCCGAA CGGGUUAUCG UUGCAAAGAU AUCCGAAGAA CUAUGGUAAG C GAGUGAAA GACAACACUG ACUAGGAUAG CUGGUUUUCU GCGAAACCUA UAAUAGUAGG CAAUUUAAGU AACAUCUUAG UA GGUACAG AACUUAAUCU CAGACAAGAU GUAGAUUUUC AUACCUAUGU UUAGGUAUGA AAUGCAUUUU UUUUUGUAUA CAU CGGGGG AUCGUGAAGA UUUUAUCGGU GAGUAUGUAG ACUCGGAAUG ACAAAGAUGA AUCUUGAAUA AUCAGACAUA GAAU GAUAA GGUUGUAUGU CAAAAGGGAA ACAGCCCAGA ACAAGAGUUA AGGUUCCAAA AUUAUUAUUA AGUGAAAUAA AGAAA GUUU UUAUAUAAGU CGACAAGAAG AUGGGCUUGG AAGCAGCCAU AAUUUAAAGA UCUCGUAACA GAGCACUUGU UAAAUC UUA AAAGCAUCGA AAAUUUAACG GAUCUAAAUA AUAUACCGAA ACCUUGUCCA UAUGUAACAU UAGUAAUAAU AUGCUAU UA AUGUUAUUUG AUGGGGUAGC AGAACGUUGA GUGAAUCUUA GAUUUUUUUU UUAUAACUAA AUAUAGAUGA UAACUCAA G UGAGAAUGGU GACAUGAGUA ACAAAAAAGA GUUUAAGGUA CCUAAAAGGU AUCUUAGAGU CUCGCCUAAA GCUUAUGGC UACGUCAAGU AACGGCCUCU AAGUUUAUAA UCUGAAGAUU AUGACGAUGA GAAAAUAACG CGCAGAAGUG CGCUGCUUUG AUACUUAUG GUACCAACAU UUAAAAGUGA AAAUUGUGCA GGAAGGAUCA GUAUCCUUUC AUUCUUAUGU GGGGGAGUGG A CAAAACUG AACAGAGUGU AUCUGAACAC AGAUGAGUCC ACACCCCCCC CCAUGUAAUG AAUGAAUGAC AAACCGUACC UA GAAUCUG AAACAAGUAA GCUAGUAGAG AAUACGAAGG CGUGAAUGAG AUAACAAUCA UAAAGGAACU CGGCAAACUA ACU ACCGUA ACUUAGGGAU AAGGAGAGCU CAUUAGUCUC GAUUAAUACG AGUAAAAAGG AAGAAGCAUG GAAUAUUGUU GUAC GACUG UUUAAUUAAA ACAAAGCACU UUGCAAAAAG ACGAUAAGUC UAAGUAUUGA GUGUGAUUUC UGCCCGAUGC CGGCU GGUU AACGAAUUUU CUAAAUUGAA AAAAAAUUUG GUUUCAGAGG AACCCCCGGU UAAUGGCGGC CUUAGCGUGA GGGUCC UAA GGUAGCGAAA UGCCUUGGCC GUUAAAUGCG GUCUUGCAUG AAUGAUGUAA CGAUACAACA GCUGUCUCUA UGAUUGA CU CAGUGAAAUU GGAAUAACUG UGCAGAUACA GUUUACCUCU AGUUAGACGA GAAGACCCUA UGCAGCUUUA CUGUUACU A AUUAUUGAAU ACGAUUCUGA AAAUUUCCAG UGUAAAAGGU AAUCGAUAAG AUAUAAUUGA AACACCUUUA UUUUUCUAU CGUAUUAUUA AACCUUAAAU UAAGGAACAA UUGUUAGAAG ACAGUUUAUG CGGGGCACAG GCCCCAUAAA GAGUAAAUGG GUGUGUCUA AAAUUUAUAA AUUUAUGUUU GCAAUUUUUU AUAGUGAUUA UAUAUCAAAU CAUCUUUAUG CUAUUCAUAG A GUGUAUUU AUUAUAUUCC UUGGGUACAG UAUAAAAAUU AUAUAUGUAU UAAUUUACAU AUAUUUUUUC UAAGAAAUUA GG UAAGAUU UUGUUUAUAG AGAAAUUAGA UGUAAAAAAA AAAUCUUAUG AGGGCGGUAU UUAAUAAUCC GCUUCUAAUA UUU UUUUGU AGUUAUUAUU AUAAAUUUAA UAAUAAUCAU GUUUAUUACU UAAAAAGCUU AAUGGCUUAA UCUUGCCUUA CUGU UUGAU UAACAACAAA UCUUACAGUC GCGUAAGCGG GGCAUAGGAU CACAAGAUAC AAAAAGGAAA GAUCUUGGAU UUUUG GAAA AGCUACGCUA GGGAUAACAG GCUAAUUUGC GCAAGAGUGU ACAAAAUGAG UGCGCGGUUU GGCACCUCGA UGUCGG CUU GACUAAUCCU CAUGGAUGCA GAAACUAUGU AGGGUACGAC UGUUCGUCGA UUAAAAAGUU ACAUGAGCUG GGUUAAA UA CGUCGUGAGA CAGUAUGGUU UCUAUCUUCU AGAGGGAAUU AGAAUAUAAU AAGGAUUAAC CUUUGUACGA AAGGAACA U GGGGUACUAU UGUUAUACCU AGUUGUAUAA CAGUUUUAUU AACCUCUGGU UUACCUGUUG UUUAUGUGCC UUAUAUUAA UUUCAUGUGU GAUGCUCCGC AAGGAUAUUA CAGGGAUGUU ACCGUCACUU GAGUAAAUAC AAUAGCAUAA GCAUGGCAGG AAAGCUAAG UUAGUCAAAA AUAAGUGCUG AAAGCAUAUA GGCACGAAAU UUACCUUAAG AUAUUUCUUA AAUAUACGUA A GAAAAUAU UACGUUAAUA GGCUUAGUUU GUAAUAAUCU AGAGAUUUUA AGGAACUAAG UACUAAUUUU AUAAAAAACU GA AUGAUUA AUAUAUCUUA CAUUUUC |
+Macromolecule #78: 16S rRNA
| Macromolecule | Name: 16S rRNA / type: rna / ID: 78 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 600.339375 KDa |
| Sequence | String: GAUGUAAUAA AAAAAAUUUU UUUUAAUUUU AUAUUACAUC AAUAAAAAUA GAUGAGUUUG GUGAUGGCUC UGAUUGAACA CUGUCCAAA UACUUGACAC AUGCUAAUCG AACGUUUAAU UUUGGCCUAA GAAAGGGGUU UCAUCGUGGC UUAAGCUAAG G GGUUUAUU ...String: GAUGUAAUAA AAAAAAUUUU UUUUAAUUUU AUAUUACAUC AAUAAAAAUA GAUGAGUUUG GUGAUGGCUC UGAUUGAACA CUGUCCAAA UACUUGACAC AUGCUAAUCG AACGUUUAAU UUUGGCCUAA GAAAGGGGUU UCAUCGUGGC UUAAGCUAAG G GGUUUAUU GUGGCUUAAG CUAAGGUUUA AUCUUUGACU UAAGCGGGUG UUUUAGGGGA ACUUGUGCCC CUAAAACCUC UU AAUUAAA AGUGGUGUAC AGGUGAGUAU AAUAUUUUUU CGCUUAACUU AAAGUGAAGG CAAAUCCUUC AUAUUGCAAA AGG AUAUCU UAGGCACCUG UUGAAAGGGG CCUACUUAUA UUAUAUCCGC UUUAAGAGGA UGAGAAAAGU UUCAGAGAUA GGUA GUUGU UAAGGUCAUG GCUUAACAAG CCAAUAAUUC UCUUAGUCGA AGCUGAAAAG GCUGAUCGAC CACAUUGGGA AUGAA AAAA UCCCAAGGCA AAUAGGUACA GCAGUGAGGA AUCUUGGUCA AUGGGCCCAC GCCUGAACUG GUAACUUGGA GGAAUG AGG GGUCAACUUU GCAAAUGGAU GAGUGAUCGU UAGAAGAUCC UUAGUCCCCU GGUCUUCUUG ACACAUGAGG UAUAUAC UU CUAGUCCAUA UUGGGGGGAG ACUCCACGUC GAUUUAUCGA GUAAAAUUCU GUAUACAUAU UGAUAAUGAC AAUAUGUA C AUUUGUCUUG ACUAAUUACG UGCCAGCAGU CGCGGCAAUA CGUAAGAGAC UAGUGUUAAU CAUCAUAAAU AGGUUUAAA GGGUACUCAG ACGGAAAAAU UCGCCCAAAU AUAGGGGACA AUUUUUCUAG AGUUUUAUGU AAGAAGGUCG UACUCUAGAG UGGAGAGAU AAAAUUCUGU GAUACCUAGG GGACGGGUAA AGGCGAAGGC AAUCUUUUAU GUAAAAACUG ACGUCGAAGG A CGAAGGCA AAGGGAACAA AAAGGAUUAG AUACCCCAGU AGUCUUUGCA GACAAUUAUG AAUGCCAUAG GUUAGAUUUU UA AUUUAGU CUAUAAAUGA AAGUGUAAGC AUUUCACCUC AAGAGUAAGG CGGCAACGCA GGAACUGAAA UCACUAGACC GUU UCUGAC ACCAGCAAUG AAGUAUGUUA UUUAAUUCGG UGACCCACGA AAAACCUUAC CACAAUUUGA AUAUUAAUAA UAAU GAUAU UAUUUUUUAU GCUUGAUAUG GCAAGCACUC AAUUUUCCCC UCCCCGUAGG UUUGCCGCGG GGGGGGAGAA AAAAG AAAA AUAAUGGAUA AUAUAGUAAA UACCAUAUUC CAACUAUAUU UAAUUAUUAA UACAAGUGUU GCACGGCUGU CUUCAG UUG AUGUUGCGAA ACUGUGGUUC GUUCCAUGGA AUUAACGUAA ACCCUUGCUU UAUUUGUAAA UAUUAUAAAG CAGUUCA CC UUUAUAUAGG AAAUGAUAAA AGGGAUCAAG ACAAGUCAUC AUGGCCUAAA UAUUGUGGGC UAUAGACGUG CCACAUUU U CCUAAACAAA GAGAUGCAAA AAUGUGAAUU UUAGCUAAUC UCAAAAAAUA GGAUAAAAAU AUACAAGGAU UGUAGUCUG AAAUUCGACU GCAUGAAUAA GAAAUUGCUA GUAAUCGUGA AUCACCAUGA CACGGUGAAU AUUCCCUCGG AUUGGUACUA ACCACUCGU CACAUGCUGA AAGGAGUGCG UGCAAUAAGU UUGCUUUUCU GUUAUAAGUA AGUAGACAUA UAGGUUUAGA U GUUAUAAU AGGAUCCUUC GUAUGCGCGG CUCUGAUUAG UGUUAAGUCG AAAUACGGUU CGUGUAGUGG AAGUUGCACG GG ACUUAUC AAUGUUGAAC AAUACGA |
+Macromolecule #79: P-site-tRNA
| Macromolecule | Name: P-site-tRNA / type: rna / ID: 79 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 23.481033 KDa |
| Sequence | String: GGCUGAGUAG CUCAAAGUAG AGCAUACCGU UCAAAACGGU AAUGAAUGUG UUCGAUCCAC AGCUCAGCCA CCA |
+Macromolecule #80: mRNA
| Macromolecule | Name: mRNA / type: rna / ID: 80 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 3.431026 KDa |
| Sequence | String: AAUAUUUUUG U |
+Macromolecule #2: 60S ribosomal protein L2, mitochondrial
| Macromolecule | Name: 60S ribosomal protein L2, mitochondrial / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 42.260766 KDa |
| Sequence | String: MLQPQFRPLL AGTASFAQTV LGRTVARCYA TKAATQSASS TSTSNTSKDA KAKIVTPYVR DAGMMRTYKP HTPGIRHLKR PINDHLWKG RPYLPLTFPK KGQSKGGRNH SGRVTVRHRG GGHKRRIRMV DFERWIPGPH TVLRIEYDPG RSAHIALVKE E ATGRKSYI ...String: MLQPQFRPLL AGTASFAQTV LGRTVARCYA TKAATQSASS TSTSNTSKDA KAKIVTPYVR DAGMMRTYKP HTPGIRHLKR PINDHLWKG RPYLPLTFPK KGQSKGGRNH SGRVTVRHRG GGHKRRIRMV DFERWIPGPH TVLRIEYDPG RSAHIALVKE E ATGRKSYI VAADGMRAGD VVQSYRSGLP QDLLDSMGGV VDPGILAART CWRGNCLPVS MIPVGTQIYC VGSRPDGKAV FC RSAGTYA TIISKEEETR EDGTKVMTGK FVNVRLQSGE IRRVSKDACA TVGIASNIMH HYRQLGKAGR SRWLNIRPTV RGL AMNAND HPHGGGRGKS KGNRHPVSPW GTPAKGGYKT RRKSNVNKWV VTPRVRNMGV RRNKKTT UniProtKB: 60S ribosomal protein L2, mitochondrial |
+Macromolecule #3: Related to ribosomal protein L3, mitochondrial
| Macromolecule | Name: Related to ribosomal protein L3, mitochondrial / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 41.262527 KDa |
| Sequence | String: MAPRLPARCW RQLSLVERSA THTTTTAAAS SLLLAGRTTP TFSASSPSTV FPSLLPQITK RGVKYGWSTL PKRSRPTRFN QVTQGLPAP TSGPAAALKR REKTTPLRTG VLAVKKGMTV FMGRTGARIP CTVLQLDRVQ VVANKTRAKN GYWAVQVGLG E RRAENVGA ...String: MAPRLPARCW RQLSLVERSA THTTTTAAAS SLLLAGRTTP TFSASSPSTV FPSLLPQITK RGVKYGWSTL PKRSRPTRFN QVTQGLPAP TSGPAAALKR REKTTPLRTG VLAVKKGMTV FMGRTGARIP CTVLQLDRVQ VVANKTRAKN GYWAVQVGLG E RRAENVGA PQLGYYEAKG IPPKQTLAEF KVRNQDGLLP VGVQLFPDWF HVGQVVDVRG ITRGMGFAGG MKRHGFAGQE AS HGNSLNH RTIGSVGGSQ GSGSRVLPGK KMPGRMGAQQ HTVQNLPILM VDNELGIVVV KGAVAGHKGA VVKVQDAVKK APP PEEFVE ATKQLLNERF PDAEEKLQAA RKLHLELKEA RRQGLIDSLI KNGLTEKADG NAAVEASA UniProtKB: Large ribosomal subunit protein uL3m |
+Macromolecule #4: 60S ribosomal protein L4, variant
| Macromolecule | Name: 60S ribosomal protein L4, variant / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 36.402785 KDa |
| Sequence | String: MAGKGLKSLN EAMNALSIAS KSCRALPMRQ SSILPCRRSM ASVATPPAAN ITRSVSEPWQ PITNVPVTVY SFPELEPRSL ESYSARHLH LPLRRDILHL AVIYEGDSTR RGMASTKTRY EVHGSHKKMS PQKGTGNARR GTRQSPLMKG GGKTFGPKPR D FSTKLNKK ...String: MAGKGLKSLN EAMNALSIAS KSCRALPMRQ SSILPCRRSM ASVATPPAAN ITRSVSEPWQ PITNVPVTVY SFPELEPRSL ESYSARHLH LPLRRDILHL AVIYEGDSTR RGMASTKTRY EVHGSHKKMS PQKGTGNARR GTRQSPLMKG GGKTFGPKPR D FSTKLNKK VYDLAWRTAL SYRYKRGELI VTEDGLDLPL PNDFLWLAGG GKLSRELEDG YVRKWVHEFM TSLNWGKEAG RT TFITGDK RPNLFTGFEL AGAEGRALEL WDVDVKDLLE TGRIVIERSA LKEMIEDHQS DLVTRVAVQG LRQKGPNLGE VLV RAPRY UniProtKB: Large ribosomal subunit protein uL4m |
+Macromolecule #5: Related to ribosomal protein L5, mitochondrial
| Macromolecule | Name: Related to ribosomal protein L5, mitochondrial / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 39.170875 KDa |
| Sequence | String: MASLRGVSRS ARALQPFSAQ FAVRRCASTQ TGAGAAAATP KSNIPDLAEL ETRSALDAPI PSEEDKKEFR PWKRAADRKA RLPSSRYQY HPPKYNRGPL HPIQSPPSSD PIARDFVPGP FNMPRLKETF RTVMASDLMT LAYIHTPPGT PKKEPTERLR A WEGDSPYF ...String: MASLRGVSRS ARALQPFSAQ FAVRRCASTQ TGAGAAAATP KSNIPDLAEL ETRSALDAPI PSEEDKKEFR PWKRAADRKA RLPSSRYQY HPPKYNRGPL HPIQSPPSSD PIARDFVPGP FNMPRLKETF RTVMASDLMT LAYIHTPPGT PKKEPTERLR A WEGDSPYF ANRARRAPRG APELPIRERD ISFRNIPEIK EITVSTFVPL GLKNPDLLIV ARAVLLAMTG TMPEMTRSKN NV VQWQLQA NKPAGCKTTI YGNAAWEFMD RLIHLVLPRI KDWKGVPAST GDGSGNVQFG LNPEDVQLFP EVECNYDMYP AKM IPGCHI AIKTTATSDR QAKLLLQSLG VPFYSN UniProtKB: Large ribosomal subunit protein uL5m |
+Macromolecule #6: uL6m
| Macromolecule | Name: uL6m / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 28.629158 KDa |
| Sequence | String: MFAPSRRRVL EAVASSSSSP VVTLPGFLVP AFQQTAGAAA RRNFSATTTR PSKLGRTPLS IPPGVEITIG EPFVKRDMTQ WKQQPKRKI TVQGPLGQLE MDIPDFIKID HDAEARRATL SVANRDEKEQ REMWGTTWAY LNRFIMGVSE GHTAVLRLVG I GYRATIDT ...String: MFAPSRRRVL EAVASSSSSP VVTLPGFLVP AFQQTAGAAA RRNFSATTTR PSKLGRTPLS IPPGVEITIG EPFVKRDMTQ WKQQPKRKI TVQGPLGQLE MDIPDFIKID HDAEARRATL SVANRDEKEQ REMWGTTWAY LNRFIMGVSE GHTAVLRLVG I GYRATIDT RPEKEEYPGQ QFVCLKLGFS HPVEMGVPKG MKASTPQPTR ILLEGINREQ VMTFAADIRR WRVPEPYKGK GI FVNGETI KLKQKKIK UniProtKB: Ribosomal_L6 domain-containing protein |
+Macromolecule #7: RIBOSOMAL_L9 domain-containing protein
| Macromolecule | Name: RIBOSOMAL_L9 domain-containing protein / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 33.173176 KDa |
| Sequence | String: MTASALSKWP TCLACLRRLA QPFGTSAARH GEPRARAVPV IRPIVHTQTR AASHRMRLQD QGVVVRLLED IPKFGRKHAI FRIERGRMR NEWFPKNKAE YMTPARFQEL GLTRDAIGEV DRSFVIMSAL EAATRPKPEE QKTEEPVPEV QISQVNVPDV T PETAHALL ...String: MTASALSKWP TCLACLRRLA QPFGTSAARH GEPRARAVPV IRPIVHTQTR AASHRMRLQD QGVVVRLLED IPKFGRKHAI FRIERGRMR NEWFPKNKAE YMTPARFQEL GLTRDAIGEV DRSFVIMSAL EAATRPKPEE QKTEEPVPEV QISQVNVPDV T PETAHALL SELIPNTLTF HREPVPIPIS QPKPALEPKI SPLIARHVPA STPETPSTGE ARRAIFGSVS SSDILNQIKA LV SGHEEAS RIVLGPSSVK IVGLAEDNDR IRHLGRWEIE IAVARAGGLD PVRKSVEILP SAQ UniProtKB: RIBOSOMAL_L9 domain-containing protein |
+Macromolecule #8: Related to ribosomal protein YmL11, mitochondrial
| Macromolecule | Name: Related to ribosomal protein YmL11, mitochondrial / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 37.363766 KDa |
| Sequence | String: MSLRLSRPAV RGLGSAIKSS RISSRSAALL VPSTSTSAFS TASPQRAAAA GHLRLPDDYV PPTQPPSARP VDTRKSQLLR TYTSMLRST PLMLIFQHNN LTAIEWAAIR RELSLALSNV PVPEGAPDIT SKIHLQVVRT RIFDVALKTV EFFDPSTVEP T TATTATGT ...String: MSLRLSRPAV RGLGSAIKSS RISSRSAALL VPSTSTSAFS TASPQRAAAA GHLRLPDDYV PPTQPPSARP VDTRKSQLLR TYTSMLRST PLMLIFQHNN LTAIEWAAIR RELSLALSNV PVPEGAPDIT SKIHLQVVRT RIFDVALKTV EFFDPSTVEP T TATTATGT KVPATYNHDL SKHAWKAVKE ATKNTEAVEK TVYGQLAPLL VGPVAILTLP SVSPAHLGAA LSVLAPSPPA FP APSRKKN PGYYDLTCQS GLQKLLLVGG RIEGKAFDYD GIKWVGGIEN GIEGLRAQLV HMLQSAGMGL TSVLEGAGKS LWL TMESRR SVLEEEQNPK KEGEGEEEKK E UniProtKB: Related to ribosomal protein YmL11, mitochondrial |
+Macromolecule #9: uL11m
| Macromolecule | Name: uL11m / type: protein_or_peptide / ID: 9 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 16.629375 KDa |
| Sequence | String: MSKAAKGGAG GVEQVVKLIV GAGQASPSPP VGPALGSKGI KSMDFCKEFN ARTAHINTGT PMPVRVTVRP DRSFHFDVRT PHTSWLLLN AAEAPIGKGG KRKGASNPGK EVVGTVSLKH VYEIAKIKQS ELRLSGLPLE GLCRAVIYQA RSIGINVIP UniProtKB: Mitochondrial 54S ribosomal protein YmL19 |
+Macromolecule #10: uL13m
| Macromolecule | Name: uL13m / type: protein_or_peptide / ID: 10 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 20.334576 KDa |
| Sequence | String: MSQTVGATRL AYSRVWHHIS AVTPHPTLST IKAPSEAITP PSLGRLASRI ATILMGKHKP IWDPSTDCGD YVVVTNCAGL YTTGHKKWR KTYYRHNTRP GSLQAITMDA LMEKHGGAEV LRKAVSGMLP KNRLRDKRLA RLKAFEGDAH PYKENLVRFG G KVVGAPGW EEAVKAIREA DMERL UniProtKB: Ribosomal protein L13 |
+Macromolecule #11: Ribosomal protein L14
| Macromolecule | Name: Ribosomal protein L14 / type: protein_or_peptide / ID: 11 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 14.13563 KDa |
| Sequence | String: MIQLKTMLNC IDNSGAALVE CAMVVGQKRH ASIGDRIVVV VQKQRGADSA GMAASSAATK VKRGDIRHAV VVRTKQKVQR RDGSVVRFD DNACVLINKA GDPIGSRING VVGQELRKKK WSKILSMAPM QA UniProtKB: Ribosomal protein L14 |
+Macromolecule #12: Ribosomal protein L15
| Macromolecule | Name: Ribosomal protein L15 / type: protein_or_peptide / ID: 12 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 33.951125 KDa |
| Sequence | String: MPPRLPLAQA ARCCQASLTA RPSVPASSPT SSLISLFAAL SVQTRSASIL ASLSDNRGAY HKRIRKGRGP SSGYGKTAGR GTKGQKAHG HVKPWFQGGQ TPLIVSHGRK GFVNQFAADM SELNLEKLQE WIEAGRIDPT KPITPKEIIK SGIIGSSIKD G IKLLGRGK ...String: MPPRLPLAQA ARCCQASLTA RPSVPASSPT SSLISLFAAL SVQTRSASIL ASLSDNRGAY HKRIRKGRGP SSGYGKTAGR GTKGQKAHG HVKPWFQGGQ TPLIVSHGRK GFVNQFAADM SELNLEKLQE WIEAGRIDPT KPITPKEIIK SGIIGSSIKD G IKLLGRGK ESFKIPVTIT VSRASASAIE AIEAAGGKIV TRFYTKESLK RLVEGKSLHT DKPLPVGKEH VEEILAQARS LK KKYYRLP DPTSRWDIEY YRDPAHRGYL SHQLAPGESP SLYFKVPTGG EKVKVVRADK VKASKTGDVA SEKLF UniProtKB: Ribosomal protein L15 |
+Macromolecule #13: Related to ribosomal protein L16, mitochondrial
| Macromolecule | Name: Related to ribosomal protein L16, mitochondrial / type: protein_or_peptide / ID: 13 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 27.443828 KDa |
| Sequence | String: MKHNASSALL SAFQGLRISS SATPFRAASL ATSAVRRPIA PTPVSVASHV RLFSATAIQA GSWLEPNLNR KKKMMKGRPR VPTGGSTKG TTVVWGDYGL RMRDHHRRIS AQQLKLAEDT IKQRLRGQKY RLYKRVACNV GVYVSGNEMR MGKGKGSFDH W ATRVAVNQ ...String: MKHNASSALL SAFQGLRISS SATPFRAASL ATSAVRRPIA PTPVSVASHV RLFSATAIQA GSWLEPNLNR KKKMMKGRPR VPTGGSTKG TTVVWGDYGL RMRDHHRRIS AQQLKLAEDT IKQRLRGQKY RLYKRVACNV GVYVSGNEMR MGKGKGSFDH W ATRVAVNQ IIFEIRGQLH EQVIRDAFRL AGHKLPGLYE FVKKGDPPVV GITKLEDGLT VEDLKNPRKK LLMPEITQSA AS TSSTAAP PS UniProtKB: Related to ribosomal protein L16, mitochondrial |
+Macromolecule #14: uL17m
| Macromolecule | Name: uL17m / type: protein_or_peptide / ID: 14 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 22.181545 KDa |
| Sequence | String: MAGAHVKYRH LSRSSAHRQA LLRNLVTSLI KNETIHTTYP KAKEAQRLAE KLITLSKRDT ETARRSAQGI LYTPDQLLPK LFGEIRERY ANRPGGYTRV LRTEPKDQYS QAPSAILELV DGPKDMRFAM TAATVARDRE QKTESTDLTK KNMDKVTRFR K DGMRAFED ...String: MAGAHVKYRH LSRSSAHRQA LLRNLVTSLI KNETIHTTYP KAKEAQRLAE KLITLSKRDT ETARRSAQGI LYTPDQLLPK LFGEIRERY ANRPGGYTRV LRTEPKDQYS QAPSAILELV DGPKDMRFAM TAATVARDRE QKTESTDLTK KNMDKVTRFR K DGMRAFED MVASLRDMKF TAGAINPKEW KKLDR UniProtKB: Ribosomal protein L17 |
+Macromolecule #15: bL19m
| Macromolecule | Name: bL19m / type: protein_or_peptide / ID: 15 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 28.235305 KDa |
| Sequence | String: MNVTASLSRR PLGCLKAGLC QSLIRGYATA VAASTTQSTG ALPDHNFFRI ADKHTKKTRT AFAVFTPPKS PTTLPPSSVV TKSKAFKIA NSKLPPLMTK PPSDPMPLLT QQQIARMDPT GARTALFSKA RHAARVGDVL MVTHRRGGEP FAGVCLAIRR S GIDTAILL ...String: MNVTASLSRR PLGCLKAGLC QSLIRGYATA VAASTTQSTG ALPDHNFFRI ADKHTKKTRT AFAVFTPPKS PTTLPPSSVV TKSKAFKIA NSKLPPLMTK PPSDPMPLLT QQQIARMDPT GARTALFSKA RHAARVGDVL MVTHRRGGEP FAGVCLAIRR S GIDTAILL RNHLGKVGVE MWYKIYNKNV AGIEIIKRRA KRARRAKLMY MRQPKHDMGS VEQYVFAWKK MRKVLSSKGL TG GVGGGGG KQKGQESKKK N UniProtKB: 54S ribosomal protein IMG1, mitochondrial |
+Macromolecule #16: Related to ribosomal protein YmL49, mitochondrial
| Macromolecule | Name: Related to ribosomal protein YmL49, mitochondrial / type: protein_or_peptide / ID: 16 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 23.953027 KDa |
| Sequence | String: MSRALLRSVL ELRTPVTRLP PSFLLPFRPA ARFLHQTAQQ VEPTSQSDAA AAAATLLKAS PPKVTTATTP EAPAAVPTST PFSQQPPIS QQVKELLPVL AAQPGHYTTI HIHGKPYLVT EGDTVKLPFK MPGVAPGDVL RLNRASVIGS RDLTLQGAPY V DERLFECR ...String: MSRALLRSVL ELRTPVTRLP PSFLLPFRPA ARFLHQTAQQ VEPTSQSDAA AAAATLLKAS PPKVTTATTP EAPAAVPTST PFSQQPPIS QQVKELLPVL AAQPGHYTTI HIHGKPYLVT EGDTVKLPFK MPGVAPGDVL RLNRASVIGS RDLTLQGAPY V DERLFECR AIVMGTESEP MRVMIKKKRR CRKKKHVFSK HKYTVLRINQ LKINDVSSL UniProtKB: Large ribosomal subunit protein bL21m |
+Macromolecule #17: Mitochondrial large ribosomal subunit
| Macromolecule | Name: Mitochondrial large ribosomal subunit / type: protein_or_peptide / ID: 17 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 41.565996 KDa |
| Sequence | String: MSLNVPSRRL LKSAPSSANA LVPALQSLSI NTRSIHTTPS QQWSLFGWGK QKVEERNKVK EELKQSELAR TRKPTQEEIF ERVRGRLEG DSIFADTTPS SQLAAEQDKH KDWATASAEK KLAAAHAKDQ TPTGISLKKD HLVRVVDPDP RSRVRWERKM V IRKLQRGT ...String: MSLNVPSRRL LKSAPSSANA LVPALQSLSI NTRSIHTTPS QQWSLFGWGK QKVEERNKVK EELKQSELAR TRKPTQEEIF ERVRGRLEG DSIFADTTPS SQLAAEQDKH KDWATASAEK KLAAAHAKDQ TPTGISLKKD HLVRVVDPDP RSRVRWERKM V IRKLQRGT DPWSVEPKAE RIARTERKLV YKTGYLPTSV KKLVHLSRQI RGKTVSEALV QMQFSKKKMA KEVKTELLRA EA KAIVTRG MGLGKAAAAA AQKETGAEPV KIQTKDGKHL EIRDPTRIYV AETFVNKGFT RGVELDYRAR GRVFKMNKPT TTM TVVLKE EKTRIREHQE RVAKKLRQGP WVHLPDRPVT SQRQFYSW UniProtKB: Large ribosomal subunit protein uL22m |
+Macromolecule #18: uL23m
| Macromolecule | Name: uL23m / type: protein_or_peptide / ID: 18 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 26.005465 KDa |
| Sequence | String: MASVAKTGAE ALRNAPFRVG KKQIFLPNHV ITFVRPLPKQ PPNLATFIVP LEFNKLDLRD YLYHVYNVEV TGVRSFVNQR MHEQRHGDV GNWRRPKSQK MMIAELAKPF VWPKPPAEDA REAFDYAMWK KQKTAQEQDT KFQGIRAQGE VVPPSQFEVT K DRKAIKAR ...String: MASVAKTGAE ALRNAPFRVG KKQIFLPNHV ITFVRPLPKQ PPNLATFIVP LEFNKLDLRD YLYHVYNVEV TGVRSFVNQR MHEQRHGDV GNWRRPKSQK MMIAELAKPF VWPKPPAEDA REAFDYAMWK KQKTAQEQDT KFQGIRAQGE VVPPSQFEVT K DRKAIKAR QSKFLSGEET WAPGKVNAFL NSKIVPEEEG WSEVEENLPL DESAESAAEE SSSKGSETRQ UniProtKB: Mitochondrial ribosomal protein subunit L23 |
+Macromolecule #19: KOW domain-containing protein
| Macromolecule | Name: KOW domain-containing protein / type: protein_or_peptide / ID: 19 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 45.460117 KDa |
| Sequence | String: MDKLLRRVRM AEGMVARRAQ RKNALLKRIT ERKQNKKNGE AFTEAIQQRK AAVEARNEDW MLGPLAPRRE LDEITLSNGN FFGSLSPTR ALLESEVSEE ERKARVAWCG SPKFLCIAPG DRVVVIEGHH KDLIGTIEKL NTRNMTVEIQ SEKLKTNTTV P QFMQNDAD ...String: MDKLLRRVRM AEGMVARRAQ RKNALLKRIT ERKQNKKNGE AFTEAIQQRK AAVEARNEDW MLGPLAPRRE LDEITLSNGN FFGSLSPTR ALLESEVSEE ERKARVAWCG SPKFLCIAPG DRVVVIEGHH KDLIGTIEKL NTRNMTVEIQ SEKLKTNTTV P QFMQNDAD KPVTQIYARL PISSVRLVHP LKDPQTGEYR DVIIRELRPR NIVHDRPTRT RSMRRFVPGE NIIIPWPKQE PI KREDQPA DTLRIDVDEK TFVPTLFRPP APQQVLDELR NKYSIFRTRH TPEYIAKKEQ EEQEKEAKKS AAKAMLTPVQ EYN RKQREL RRARGQPALT EEMLAKIGEV VARNKLGHHQ APKVKEETVA QIEKAVEQLS LGGGQEDAAT TTSPEQPKVV UniProtKB: KOW domain-containing protein |
+Macromolecule #20: Related to 60s ribosomal protein L2 (Mitochondrial)
| Macromolecule | Name: Related to 60s ribosomal protein L2 (Mitochondrial) / type: protein_or_peptide / ID: 20 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 49.549199 KDa |
| Sequence | String: MHLAQLRQPL TRAAANSCHS VCRLPTTHSS VSATAILSEQ FAHLRIGPSS SNVAVEGRRY ASVKAQGAYR LKPKRTIPKK LGAKKTGDQ YVIPGNIIYK QRGTLWHPGE NTIMGRDHTI HAAVAGYVKY YRDPQLHPDR QYIGVVFNRN DKLPYPKDAP R KRKLGLVA ...String: MHLAQLRQPL TRAAANSCHS VCRLPTTHSS VSATAILSEQ FAHLRIGPSS SNVAVEGRRY ASVKAQGAYR LKPKRTIPKK LGAKKTGDQ YVIPGNIIYK QRGTLWHPGE NTIMGRDHTI HAAVAGYVKY YRDPQLHPDR QYIGVVFNRN DKLPYPKDAP R KRKLGLVA VPRKVEEVEK PTMSASGLPL FVTRHETIEI PPPAVTTPAA AGKAVKGQGA RVSASGAAAV PASSSTISAS AS SNSNNGG SSVIAELIKE KLAARAEYNA RQSALRKLQQ QKMLARRGTR VLRLMNNYSY RETNWEIGRL IGDPGSVPGT EKV GSRKAK FRARRRRRNT FLLGIKERKL AKADRREEYR RRVREKREQR LVQRKEFLAK QREAKKAREG GAAAEKSEKK EVKA EKPAA AAPKVEKPKA PEAAKKESKP KVEEKKAAAA EPKKDSKTEK KD UniProtKB: Large ribosomal subunit protein bL27m |
+Macromolecule #21: bL28m
| Macromolecule | Name: bL28m / type: protein_or_peptide / ID: 21 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 31.501131 KDa |
| Sequence | String: MSSFLVRPFC LRATTTTTTT TTTTALVQQQ LRALTTTTAL TYHQPRVPSS AIPIPNVSGR VPLPDSTPVP EDFKIEIPSY PYGPRRVYH QSNTGLYGSA LIRFGNNVSK RNEIKTRRKW RPNVQQKRLW SKSLGVFVRT RVTTRVLRTI DKVGGLDEYL L GHKAARVK ...String: MSSFLVRPFC LRATTTTTTT TTTTALVQQQ LRALTTTTAL TYHQPRVPSS AIPIPNVSGR VPLPDSTPVP EDFKIEIPSY PYGPRRVYH QSNTGLYGSA LIRFGNNVSK RNEIKTRRKW RPNVQQKRLW SKSLGVFVRT RVTTRVLRTI DKVGGLDEYL L GHKAARVK ELGPWGWMLR WRIMQTPEIR ERFAKEREAL GLPPRREEDE EKSLEEQLRE FQVAGIQFAE GKAPKTKGQL KE EADRLIK KSETEEFGLG QEEDLFMKEE PKPTKMA UniProtKB: 54S ribosomal protein L24 |
+Macromolecule #22: 54S ribosomal protein L4, mitochondrial
| Macromolecule | Name: 54S ribosomal protein L4, mitochondrial / type: protein_or_peptide / ID: 22 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 29.64526 KDa |
| Sequence | String: MASPAALRPS MGAIMQTCRS AATAPKVAVA VPVRALSTSA ALLKRHKYPT ARVTRDNSKQ RGESALRKSG TRWKLSVSDE PLPEPVPRE ELPPIQVDEN HGLWDFFYDR ETVAMAPLEH TKHGRAWTVS ELRKKSWDDL HRLWWVCVKE RNRIATANWE R TKSELGFG ...String: MASPAALRPS MGAIMQTCRS AATAPKVAVA VPVRALSTSA ALLKRHKYPT ARVTRDNSKQ RGESALRKSG TRWKLSVSDE PLPEPVPRE ELPPIQVDEN HGLWDFFYDR ETVAMAPLEH TKHGRAWTVS ELRKKSWDDL HRLWWVCVKE RNRIATANWE R TKSELGFG LAEANERDRN VKQTMRGIKH VLTERFYAWE DAVKLAEQDP EIDLSNPENP YTPSTFLEAE ETAEGAEASE TQ STTTEID PTTIPSSKSQ AEAPRV UniProtKB: 54S ribosomal protein L4, mitochondrial |
+Macromolecule #23: Related to ribosomal protein L30
| Macromolecule | Name: Related to ribosomal protein L30 / type: protein_or_peptide / ID: 23 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 18.196393 KDa |
| Sequence | String: MSFFRITLHR SAIGLPKRTH GVLAALGLRR RNQTVFHPVE PQFAGMLMKV KELVRVEEVP HRLTKRELKD ERKPDAGFVV EKQVRRFIP GVGVQDEVDF SKVVEELKAQ KVENVEGVEK MVVEGGAVVR DGKRAGDLGV RTRSDWEVIG KMKSVLPKVK A L UniProtKB: Large ribosomal subunit protein uL30m |
+Macromolecule #24: Related to ribosomal protein YmL36, mitochondrial
| Macromolecule | Name: Related to ribosomal protein YmL36, mitochondrial / type: protein_or_peptide / ID: 24 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 23.766729 KDa |
| Sequence | String: MSSKLPTTLL RRPSALPSTT TYTAYSASRP TPPSCTAHGA QGQQIRNATF VPRHRRPYQF TQLVQLSDGS TFTVRTTMPT ALYKSAKDS RNHLLWQPSD KSLKNVELDE AGKLAAFRER YGRGWDLDAK MTPEEEAAAA AALAAGGGAG VPGGKAAKKA A EEALLAKK ...String: MSSKLPTTLL RRPSALPSTT TYTAYSASRP TPPSCTAHGA QGQQIRNATF VPRHRRPYQF TQLVQLSDGS TFTVRTTMPT ALYKSAKDS RNHLLWQPSD KSLKNVELDE AGKLAAFRER YGRGWDLDAK MTPEEEAAAA AALAAGGGAG VPGGKAAKKA A EEALLAKK KKEEEEAAKK AAEAEEADPF DSLTDLISGY ATENMNPGLN FKETRHYGKK K UniProtKB: Related to ribosomal protein YmL36, mitochondrial |
+Macromolecule #25: Related to ribosomal protein YmL32 (Mitochondrial)
| Macromolecule | Name: Related to ribosomal protein YmL32 (Mitochondrial) / type: protein_or_peptide / ID: 25 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 14.114779 KDa |
| Sequence | String: MAAMTAAAAA PAMRLFNPST FFQTLRTQRF GVPALNFAVP AAAISLLPSI PSLLEDIWEG ILRAVPKKKT SHMKKRHRQM AGKALKDVT HLNRCPACGN LKRMHHLCST CLGKLKGFMD RNGGSNAKAY UniProtKB: Large ribosomal subunit protein bL32m |
+Macromolecule #26: bL33m
| Macromolecule | Name: bL33m / type: protein_or_peptide / ID: 26 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 7.027574 KDa |
| Sequence | String: MAKKAKSRII NVRLISMAMT GYFYTFTRPR TSLPMSMLKY DPIVRRKVLF LEQKRKGRS UniProtKB: Ribosomal protein L33 |
+Macromolecule #27: Related to ribosomal protein L34, mitochondrial
| Macromolecule | Name: Related to ribosomal protein L34, mitochondrial / type: protein_or_peptide / ID: 27 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 14.853288 KDa |
| Sequence | String: MSRIFSSALQ AVFKPSAFLP KTATRSFSIL PSLRPATLSS TPSTIFRAPN AITAPSASPS ATTGIDGEVL DLLSASSLIS SHPALSGLG SQIRCGPRPT MSGATRLVQK RRHGFLSRVK TKNGQKTLKR RLAKGRLRLS A UniProtKB: Related to ribosomal protein L34, mitochondrial |
+Macromolecule #28: Ribosomal protein
| Macromolecule | Name: Ribosomal protein / type: protein_or_peptide / ID: 28 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 13.960405 KDa |
| Sequence | String: MSNLFRSLAS SMRALSLAAP RATAVNTTKT VVSTHQTRCL SQGLLSRHIC TPMCSHNRPV AVCQSAKNGL QSKQQSRGMK VHSAIKKRC EHCKVVRRKA NKRQNGYLYI ICPANPRHKQ RQGYR UniProtKB: Large ribosomal subunit protein bL36m |
+Macromolecule #29: PEBP-like protein
| Macromolecule | Name: PEBP-like protein / type: protein_or_peptide / ID: 29 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 50.749188 KDa |
| Sequence | String: MSTCQQVARP LVRCLRQTGP GAATCSTRSV AVAAVRPFSS TPMRKEEGAT TTTTTSPVDS TTTPPPLPSA TEAKATVADT VVGGPELGS RRRRAALATT GNLPFEQLPY QCFQDARKIL QQDRAAKIAQ IVKETEKIKL IEARDASEFE GGEAAKQTRI K SLRKYIEE ...String: MSTCQQVARP LVRCLRQTGP GAATCSTRSV AVAAVRPFSS TPMRKEEGAT TTTTTSPVDS TTTPPPLPSA TEAKATVADT VVGGPELGS RRRRAALATT GNLPFEQLPY QCFQDARKIL QQDRAAKIAQ IVKETEKIKL IEARDASEFE GGEAAKQTRI K SLRKYIEE LKILADINDP EVKRRFEDGR GDMTKPVYRF MAERRWRSMD YKIIAQRISQ FHVVPDLLPA FDPTMDVKLS FR GYQVSPG AILDSRVTEV APTLRMQVFD KGERLLTVVV IDSDVPDVTH DNFKRRCHFL AANIPWDPSK TVLSLRSVGD RVE GDVGKP WLPPFAQKGS PYHRLNVFVL EQKPGAKIDG EALKKHLENR ENFSLKGFRE KFDLEPVGFN LFRSEWDEGT AEVM ERHGI PGAEVEFKRQ KFASLKPPRK ARGWEAKRQK PKYKSLWKYV KRIA UniProtKB: PEBP-like protein |
+Macromolecule #30: mL40
| Macromolecule | Name: mL40 / type: protein_or_peptide / ID: 30 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 41.496336 KDa |
| Sequence | String: MVSATKKPVS AMSAKNSTRH RKVSHKVTKT IATAKSQVIS NKAMKIRTKS WKDMPSPKRR RRINAMVDEL RLQGNVKMPK PGKSTYKAG GGERVRLVKA ASTLPTPPLT PPVLSKPTGS ASQSKGDLKP KRPTPEKKRS VSKRDVERFF LSTQTSTRRR E NTPIHAPI ...String: MVSATKKPVS AMSAKNSTRH RKVSHKVTKT IATAKSQVIS NKAMKIRTKS WKDMPSPKRR RRINAMVDEL RLQGNVKMPK PGKSTYKAG GGERVRLVKA ASTLPTPPLT PPVLSKPTGS ASQSKGDLKP KRPTPEKKRS VSKRDVERFF LSTQTSTRRR E NTPIHAPI MSSTFTSLRG LATRLFAGGA RPSPSSLLLP NKPATAALPT AIQHQQTASY ASKGKSGPPA GMFSGQKAGS KK SKGPKQV DPRIINILRH FAVLSPKRIP PPLRFGRNRY LRHWTIHRAW LLFRRQQREQ RERILMQQHQ SMSNACEELR NTE GPGTRE TGYLYRVAML KNGVYGLKSI PIEYASRALV ETPGRQAWNH EWKR UniProtKB: Uncharacterized protein |
+Macromolecule #31: mL41
| Macromolecule | Name: mL41 / type: protein_or_peptide / ID: 31 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 11.796603 KDa |
| Sequence | String: MQPTRILQGL KYRKLRLTTK DVNKGFYKGN RTGSMGTHTS YGTYKIDYTK VRTYVCPDLT GFKLTPFVSK TIRPVHDQFP GDKLGPKNP ATYLARWKSE NGLD UniProtKB: 54S ribosomal protein L27, mitochondrial |
+Macromolecule #32: L51_S25_CI-B8 domain-containing protein
| Macromolecule | Name: L51_S25_CI-B8 domain-containing protein / type: protein_or_peptide / ID: 32 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 15.126693 KDa |
| Sequence | String: MTVKALTQIS SAGRNGVGAF VLQCKKLDIH YSDWAGSSRG MNGFIKSLLP KFAAANPQIE FVVSPRPAKH PILMGHYING RTKAICVRN MEPLEILKKA ELLRDASGEK PQKFKKPVTS TNPSVRGVWS PYHGQGMAV UniProtKB: L51_S25_CI-B8 domain-containing protein |
+Macromolecule #33: Ribonuclease III
| Macromolecule | Name: Ribonuclease III / type: protein_or_peptide / ID: 33 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 48.106484 KDa |
| Sequence | String: MKRIATPSLT SRLLVAARSG VSPATAAIRS RSAISVRSQS TAALAQHDAS HDLNNDRFPP LEPLPPAAES LPSPLPERAL TSAKLAALH ARLNLSPKIP LQTLARTLVD ASADENPQFN NANLAFVGQT LINYHIAEWL LCKYPRLPQG ILFSAMKAYA G PKPLLQIA ...String: MKRIATPSLT SRLLVAARSG VSPATAAIRS RSAISVRSQS TAALAQHDAS HDLNNDRFPP LEPLPPAAES LPSPLPERAL TSAKLAALH ARLNLSPKIP LQTLARTLVD ASADENPQFN NANLAFVGQT LINYHIAEWL LCKYPRLPQG ILFSAMKAYA G PKPLLQIA RSWGVDTAAV PGGEVDPGLL QFDALKPGVA ITNFGYKRTE LAYLEKFKWR RGMASRVVLD DDFGDVVRSD PK VAADLEK AMEEQDQDKT PDEEEAEMVA NEQDQDVSYD RYGNPDTRAA AERAHAYFVR AVVGAIYAHC GREAAKAFVK AHI MSRTLD IAKLFEFKYP TRELAALCAR EDFEPPVARL LSETGRQSRT PVFVVGIYSG SDKLGEGAAS SLDHARFKAA MNAL KAWYL YSPGENPRVP SDMLEEGAKP WTPAYIDMGE VISR UniProtKB: Ribonuclease III |
+Macromolecule #34: mL46
| Macromolecule | Name: mL46 / type: protein_or_peptide / ID: 34 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 41.322074 KDa |
| Sequence | String: MSASSRGAAL LRSQQRSICL QCRNQTRVLA PAGVTSAPRR FYSAEASATA TATATATTTT TLPPPHPPVT TSTGTHAATS TSSQIYRIK SGVILTRPPL LTRDLTPFEE SFYFYQKRLN ERLTAPFRKD FYFKKDTAAD LDWRIKLKER HGVPAKDIGR Y NPRGRMAW ...String: MSASSRGAAL LRSQQRSICL QCRNQTRVLA PAGVTSAPRR FYSAEASATA TATATATTTT TLPPPHPPVT TSTGTHAATS TSSQIYRIK SGVILTRPPL LTRDLTPFEE SFYFYQKRLN ERLTAPFRKD FYFKKDTAAD LDWRIKLKER HGVPAKDIGR Y NPRGRMAW NDEVLVGSQT SSRKHMVEKL LADAEMRVSE DGEEIPAEDR VPVEKPMPRR TEADEKGDVK RLDRALDKTL YL VVKKKAD KEGEEAKWMF PTGVVPTDEG LHETAARILA ESAGVNMNTW IVGRVPVAHH VVRPVFGQKD GALLKKGEKI FFL KGRIMA GQADLTDNLH DLVDFKWLTQ EELRSTLAEE YFHSVKGMFA ER UniProtKB: MRP-L46 domain-containing protein |
+Macromolecule #35: mL49
| Macromolecule | Name: mL49 / type: protein_or_peptide / ID: 35 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 18.180738 KDa |
| Sequence | String: MFRSTFFGLS RAIVQPATPL TVRAAFQSRF YSAAAAASQP TTTATTTTPL QQQQQQQPTT QPTTPIQTQT GAAPTESTKP VAKPYLVGR AWTQRLPVYH LAKRGGNKKL TQIKKVQGDG QALRRDLAQF LGLEVKEVRV KVPTGHLEVD GHRREEIVKF L DGLGF UniProtKB: Large ribosomal subunit protein mL49 |
+Macromolecule #36: mL50
| Macromolecule | Name: mL50 / type: protein_or_peptide / ID: 36 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 49.748516 KDa |
| Sequence | String: MRRIPRIPST ASALCSSSSS VASTSAAPLR TLRAAADSSS ICQSCSFSTQ TSATRNRVWQ NTVQTQRRCA STVTETTPEA PVAAAAAET ATESTETVEQ QRKRKGGFFE TQRQVNLVLP GQPTRADAPE KIRDPNYEPA TSGAGLQEIG GMSDWWSKPE H FRDGGKQF ...String: MRRIPRIPST ASALCSSSSS VASTSAAPLR TLRAAADSSS ICQSCSFSTQ TSATRNRVWQ NTVQTQRRCA STVTETTPEA PVAAAAAET ATESTETVEQ QRKRKGGFFE TQRQVNLVLP GQPTRADAPE KIRDPNYEPA TSGAGLQEIG GMSDWWSKPE H FRDGGKQF EYQGFAPQEK ITDPRLLKVI LRRALAEGLA LKKFGANPKN PADMASIIGN GDHWQRTVSV EMCRGENGEL SL KNESDLQ KVWILMRNAA EKTYYQREWQ EEINRLRSLG EKEQAKQLLE EGKKLGYRLK SEEGSLVKLT VDEAVELRKS WNN DWKEAI IRDPVVKFYA AKRIQKMTGH ILSDGKLTSI QTVANFMDAL VTPPKPKKLA EQIEQSSILP ELPNVKVYPR RVTP VDKER MVGRWKVIQK ELQKRELPVL GTGNHGKYVE LKWLGSKQ UniProtKB: Large ribosomal subunit protein mL50 |
+Macromolecule #37: Probable ribosomal protein YmL44, mitochondrial
| Macromolecule | Name: Probable ribosomal protein YmL44, mitochondrial / type: protein_or_peptide / ID: 37 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 11.128777 KDa |
| Sequence | String: MLTRFITDVS TRFNPFSAKA KAARLFLSFL PPNARSNGMN ITTQLLPRNS TETPLLYVKF KDGKEMNLDV ENMGIKSIVE EVDRHSRIL QKQADLNDG UniProtKB: Large ribosomal subunit protein mL53 |
+Macromolecule #38: mL54
| Macromolecule | Name: mL54 / type: protein_or_peptide / ID: 38 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 23.179602 KDa |
| Sequence | String: MICRTCLRRA SGFARPAAAA ATRPMVAASS RAAFSTTFSV CTPPAATPAA AAPATSTAEA GLAPVPGAAT TEQAKAAISS CPAGTKLNG LNYFKNKADP VALPDEEYPE WLWRCLEVQK KTDDAADADA GDEFSKSKKQ RRLAMKRARA QEAKILASGD L EALAPKIP ...String: MICRTCLRRA SGFARPAAAA ATRPMVAASS RAAFSTTFSV CTPPAATPAA AAPATSTAEA GLAPVPGAAT TEQAKAAISS CPAGTKLNG LNYFKNKADP VALPDEEYPE WLWRCLEVQK KTDDAADADA GDEFSKSKKQ RRLAMKRARA QEAKILASGD L EALAPKIP LQKQSVNLPG AVNGGVQDAV LADEKREELR KAMRKERKAK IKESNYLKAM UniProtKB: Mitochondrial ribosomal protein L37-domain-containing protein |
+Macromolecule #39: RNase III domain-containing protein
| Macromolecule | Name: RNase III domain-containing protein / type: protein_or_peptide / ID: 39 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 29.625195 KDa |
| Sequence | String: MAALTPSRSV FASTCKTIAQ QCSRRPVAVQ QLAAVPVARQ VSSSSAVQQE QDASGTSSSQ QPRPRWSYTP ERMKGPGFSL NLVKDPRRK QWMVNSDPAK LDAFYDAFLG QGGSRMLSEE TKWLAVTHKS FDYGRRGYNT RLAFLGRQII ALETTRSILT S PVLNEPIV ...String: MAALTPSRSV FASTCKTIAQ QCSRRPVAVQ QLAAVPVARQ VSSSSAVQQE QDASGTSSSQ QPRPRWSYTP ERMKGPGFSL NLVKDPRRK QWMVNSDPAK LDAFYDAFLG QGGSRMLSEE TKWLAVTHKS FDYGRRGYNT RLAFLGRQII ALETTRSILT S PVLNEPIV DKYGRTPYNH AALANIDKLI YTQPLDIMDK TKIARMGIDF GLLTVMRWKP RMPEDLESSG VVVVLNSTLF AI IGAISLE KGAAVAQRIV REKILKRLGA UniProtKB: RNase III domain-containing protein |
+Macromolecule #40: Related to ribosomal protein YmL20, mitochondrial
| Macromolecule | Name: Related to ribosomal protein YmL20, mitochondrial / type: protein_or_peptide / ID: 40 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 25.439018 KDa |
| Sequence | String: MEASLSLFRP VATCCRRVAL SSSSSSSQKA AAVAGISVRY QSTANRTKRM LNIPPHESFL NVPVEGDRII FNPPSSEASV YHTPFKFLP RSDPRRRANI YKLFKPPQAP ITTPESSSTD AAAADQHGDL PPVLYNPTKS YNVTPEQVEE IRELRAKDPK K YSVTYLSN ...String: MEASLSLFRP VATCCRRVAL SSSSSSSQKA AAVAGISVRY QSTANRTKRM LNIPPHESFL NVPVEGDRII FNPPSSEASV YHTPFKFLP RSDPRRRANI YKLFKPPQAP ITTPESSSTD AAAADQHGDL PPVLYNPTKS YNVTPEQVEE IRELRAKDPK K YSVTYLSN KYNCTKVFIM MCTQAPREHQ EQHKLARART AENWGPRRAA AKLDARRRKE MLHRGEI UniProtKB: Related to ribosomal protein YmL20, mitochondrial |
+Macromolecule #41: Mitoc_mL59 domain-containing protein
| Macromolecule | Name: Mitoc_mL59 domain-containing protein / type: protein_or_peptide / ID: 41 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 18.745025 KDa |
| Sequence | String: AATVKQHVQL ALSLPPRLRT FLARYPPASI LPAGADPETH KTPYQEESPN PFMPTKHPIT GKWHNPKYSL RRQAELVKLA QEHGVEELL PFTRKLNEEK LRKRVELGLR VKGTGVGEKV KGHKHERTLV AKMEKRREAM LAMPDLIREW KKVGKRNWTK F PK UniProtKB: Large ribosomal subunit protein mL59 domain-containing protein |
+Macromolecule #42: 54S ribosomal protein L31, mitochondrial
| Macromolecule | Name: 54S ribosomal protein L31, mitochondrial / type: protein_or_peptide / ID: 42 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 13.131485 KDa |
| Sequence | String: MFGAFRFTNP LSGGLLWKIP WRMSKFQKLR QRRRLRAVDN VVATIENALA KKGETVKAVE RWRAEMPTEA EMLPKDKYTI FDRKEKRYR KGIHKLPKWT RVSQRVNPPG Y UniProtKB: 54S ribosomal protein L31, mitochondrial |
+Macromolecule #43: mL67
| Macromolecule | Name: mL67 / type: protein_or_peptide / ID: 43 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 32.516111 KDa |
| Sequence | String: MNSVTTSTIG RLAFGVSRLS AVTNVAGAVR HASTHPTTAI PKPTTAVLKT SPLAEETAVE LKSPSRLQLK SEPGNKGNPK GHGDRIWAF HHIEQGQVIY STKPVISHQH LIRQQPFTGK NLVPRKIRKD YWRPLAMIEL AGKDQGAVGR SVYQKLREFK K RHELDWAN ...String: MNSVTTSTIG RLAFGVSRLS AVTNVAGAVR HASTHPTTAI PKPTTAVLKT SPLAEETAVE LKSPSRLQLK SEPGNKGNPK GHGDRIWAF HHIEQGQVIY STKPVISHQH LIRQQPFTGK NLVPRKIRKD YWRPLAMIEL AGKDQGAVGR SVYQKLREFK K RHELDWAN EAEEGRKLMH KSKRERGQEL NDQKPNSVAD MAAVLAGAGK GNLMWQVSRL GSSSELVGEI KKASGAVEKG AV VDVKRQL RRATVYWTNE QDKFHAREWS DNVSHEVGIP GEAEKKRMPI RMSSE UniProtKB: Mitochondrial large ribosomal subunit protein |
+Macromolecule #44: bS1m
| Macromolecule | Name: bS1m / type: protein_or_peptide / ID: 44 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 51.59634 KDa |
| Sequence | String: MAFSRGVSPG GALLRTSRMF SLPPVIPPPP GNKLQMISER ASATEAYPTH QVLTTFESSR SRGDWGLKRP LPLKSTTGTT YPMVKVKEM DSLEQITDFT SGTQHGLTLK KFQALNIPIS TPSEISDPSR LFRPVQRSVF EADTDVTAFS PDEQIQEAEK R WKFSGPWL ...String: MAFSRGVSPG GALLRTSRMF SLPPVIPPPP GNKLQMISER ASATEAYPTH QVLTTFESSR SRGDWGLKRP LPLKSTTGTT YPMVKVKEM DSLEQITDFT SGTQHGLTLK KFQALNIPIS TPSEISDPSR LFRPVQRSVF EADTDVTAFS PDEQIQEAEK R WKFSGPWL AGMTPGEFKE YLAKTVRPKR AEFRKFIQKK IAAQKTEAAN RELQETAALR GDAVAETQEP FKPESITDDE VT EYLRRLR NDNQVLYDLV GQFLDLAPLK PPQAAEARQH SLLVNLRATD SPYGGRGPPI THPSAGISYL RTAAYLNNHP IYG PQKSHP PVQARVLKPR RGGLGNDAKI GVAGFVADGP LGTSHSNLRG NSVMDKFDPS IEGGAKLWVN VDKATVDSTG RVQL TVSDA KATDVLIAKE LIGDAREPIF GSAPKRQEKT FKKIPMTAAR LRGRYENSDS PSSPTMSGSS GYGLR UniProtKB: 37S ribosomal protein mrp5 |
+Macromolecule #45: Mito ribosomal protein S2
| Macromolecule | Name: Mito ribosomal protein S2 / type: protein_or_peptide / ID: 45 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 47.197305 KDa |
| Sequence | String: MIVRNIGARL GRRALATPLS RASFPAQSRF LSQDNFTAPP PPPTNSKKQA AKTAFAEPTL EEQAEFYAES IEKHTPEFAA SPAAEAWSA PSATATETVT TWDPSLVDEE ALKLKQQIEA IPGNFKLFKQ TKAQTQKLGA GVEVRYIPEQ YLRNPPSDAS L EDLMAAQA ...String: MIVRNIGARL GRRALATPLS RASFPAQSRF LSQDNFTAPP PPPTNSKKQA AKTAFAEPTL EEQAEFYAES IEKHTPEFAA SPAAEAWSA PSATATETVT TWDPSLVDEE ALKLKQQIEA IPGNFKLFKQ TKAQTQKLGA GVEVRYIPEQ YLRNPPSDAS L EDLMAAQA HMGHNTSLWN PANARYIYGV RQGIHIISLE TTATHLRRAA RVVEEVAYRG GLILFVGTRP GQRPIVVRAA EL AKACHLF TKWRPGTITN REQLLGGVPL TVVDELDRPL SGFEDHLHDR RPLAPDLVVC LNPKENMTLL YECSLAKIPT IGI IDTNTN PSWVTYQIPA NDDSLRATAL ISGVLGRAGE RGQKRRLEAA QRGVVTWKTP ADVQGYFELA SARAADARRR QAQD NSVEE QKEKDTFLSE DALKAMFGGD ARI UniProtKB: Small ribosomal subunit protein uS2m |
+Macromolecule #46: Ribosomal protein S5
| Macromolecule | Name: Ribosomal protein S5 / type: protein_or_peptide / ID: 46 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 58.907285 KDa |
| Sequence | String: MNNTNTNTNN KNLASSSASV IFSKYINNNN NKLIPFKIKN SDLGRTRYFP PISKEWKNSI YVFNHNNLKN LPLFDININS LIKDYFNLQ FKDKILFKKK RLSKVKVVSL NKIYASKAEI KHTNTKAILT VYTFNREKIS LYKKIKKLKK SFYFVFDKII S FSERVILS ...String: MNNTNTNTNN KNLASSSASV IFSKYINNNN NKLIPFKIKN SDLGRTRYFP PISKEWKNSI YVFNHNNLKN LPLFDININS LIKDYFNLQ FKDKILFKKK RLSKVKVVSL NKIYASKAEI KHTNTKAILT VYTFNREKIS LYKKIKKLKK SFYFVFDKII S FSERVILS GVPIRVDKDG KVLSNLYLPF RLRGVLPWIT ESHVNIWRKI IIASLYKELI LLRKYKLRLD LNKYKFEEKL LY RLNNLIM KYYNKKVEFN IVNMRSFLLN SDILTKILAL KLKNRNARVI KIMDVILNKA NLPKINRVQE KASLIKSVDW NLL ENKFKN LNLSFILNDA SYAERNNLSE LLNKLYYNVL LVSQKGVWAL RSPKGEQPSF HSKSGGSPTK FIQKGAGAAS LGGN AKAQP KVSSFAKAKK YAKIYQIIFN SINYKNMGGL RLEIKGRLTK RYRADRSLFK VKWKGGLKNL DSSYKGLSSV NMRGY AKPN VEYSIFTSKR RIGAFAVKGW VSGK UniProtKB: Ribosomal protein S5 |
+Macromolecule #47: S4 RNA-binding domain-containing protein
| Macromolecule | Name: S4 RNA-binding domain-containing protein / type: protein_or_peptide / ID: 47 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 51.610031 KDa |
| Sequence | String: MKIRRLFKYH SLKKKPIRPT WNKYNLYNLA TAGREPRISG KTFFQQKWLA KSLTRAYHGE HIKERKWARM FSRRLPAVVN MDPAYMAKY NGSEQAAGRG SGLSEPPAYE KTADEKPAKQ VIANPGRKVP TPYEQMTFAP LERRLDIAIF RALFASSARQ A RQMVVHGA ...String: MKIRRLFKYH SLKKKPIRPT WNKYNLYNLA TAGREPRISG KTFFQQKWLA KSLTRAYHGE HIKERKWARM FSRRLPAVVN MDPAYMAKY NGSEQAAGRG SGLSEPPAYE KTADEKPAKQ VIANPGRKVP TPYEQMTFAP LERRLDIAIF RALFASSARQ A RQMVVHGA VTVNGKKMKH PGYLLNPGDL FQVDVERVLY ATGAPKDKKL LAAAMKAAKK ELAEEAQEAA EEKDAEAEEA EE GEAQEAA SKSGKKEKQV DTSKFDEAWN RKYELLQLQR LLQYAKHIAD SPSHQLSATR KQALRALKRD IKAAIKQAQR RDG SVSPTG NVVDDLTMLL ERLAVSPSKA IQQEEAAKEL SKKLDKQSRQ VLKKVIEYND EHEDNEIDPS KPYMTPWRPR NYMS AFAFI PRYLEVNQNI CAAVYLRHPV ARPGESEVPS PFSPTINQLA FNWYLRRG UniProtKB: S4 domain-containing protein |
+Macromolecule #48: Related to ribosomal protein S5 (Mitochondrial)
| Macromolecule | Name: Related to ribosomal protein S5 (Mitochondrial) / type: protein_or_peptide / ID: 48 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 53.297543 KDa |
| Sequence | String: MSAARFAARS FLSRRLAATV PAAAPAASTP AASTCPAAQQ QQSQFHSSAH LEARRRSRFK NVRAVEMGLT SDAKIESFTK KKFAEYTED EKAALAHNYP AEHMEAIEAA EAAIDPKDLT IQGRLRVDPY RMPYIDDFSE IQPIIDKRAR RSAPPSHKAR F MDVDEFTQ ...String: MSAARFAARS FLSRRLAATV PAAAPAASTP AASTCPAAQQ QQSQFHSSAH LEARRRSRFK NVRAVEMGLT SDAKIESFTK KKFAEYTED EKAALAHNYP AEHMEAIEAA EAAIDPKDLT IQGRLRVDPY RMPYIDDFSE IQPIIDKRAR RSAPPSHKAR F MDVDEFTQ DLINWADEIR RGEPTHRMKK LRDFVPEEFF EKPEGQWPKD VRDEAFTKFW AYLKEQKDAD AKAAANATGP TD GDILSYI LERSSMTDNN LQANSSLAPA LPDKVPGVEG KYRNAIDPAD DGLDDKGQYQ ELKKRTGMSV RQILQLKTKK LVH RRVVNQ TRLGKIASDS VMVIAGNGDG WLGLGMAKSV EASIAVEKAT LLAIQNMQPI PRYENRTIYG EVTTKVSGTI VRLN SRPPG FGLRVSHRIF EMCRAAGIRD LSAKFLRSRN PMNTVKATYQ ALLSQPNPED LAIGRGKKLV DVRKVYYGGS VY UniProtKB: Small ribosomal subunit protein uS5m |
+Macromolecule #49: Ribosomal protein S6
| Macromolecule | Name: Ribosomal protein S6 / type: protein_or_peptide / ID: 49 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 13.304392 KDa |
| Sequence | String: MLYETIGIVR PGNIAEVKEL VLTAGKLILN QGGVIRDIKN WGTFLLPRPI STNQQRHNRG HYFVMRYDAS IATHEEVRRT MKADPRVIR TANVKLGDGK LETLSRFGAI PWRSLEEA UniProtKB: Ribosomal protein S6 |
+Macromolecule #50: Ribosomal_S7 domain-containing protein
| Macromolecule | Name: Ribosomal_S7 domain-containing protein / type: protein_or_peptide / ID: 50 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 33.821914 KDa |
| Sequence | String: MPARLGLSAA FRSLSIRTNQ LPQQQSIAAA VQARTLITDS TTSSRLPPRV QIQQQQQQRT QPYSTETTPP PNSNNGDLAG IEGQPPVEV TPENAAALSQ LSEIAYGVKA ADVAIEGHKY GLPTLPLPSE LHHKYRYSEV VNHATKLLMK DGKLSKAQRD M AMILNYLR ...String: MPARLGLSAA FRSLSIRTNQ LPQQQSIAAA VQARTLITDS TTSSRLPPRV QIQQQQQQRT QPYSTETTPP PNSNNGDLAG IEGQPPVEV TPENAAALSQ LSEIAYGVKA ADVAIEGHKY GLPTLPLPSE LHHKYRYSEV VNHATKLLMK DGKLSKAQRD M AMILNYLR SSPPPKLNPS RPLIPGHPPA SHLPLNPVEY LTVAIDSVAP LIKMRGYKGL AGGGRSLEVP QPIPARQRRS TA FKWILDV VNKKPSKGSG RTMFAHRVAE EIVAVVEGRS SVWDKRMLIH KLGTANRANL NSPALQLKGK RF UniProtKB: Ribosomal_S7 domain-containing protein |
+Macromolecule #51: uS8m
| Macromolecule | Name: uS8m / type: protein_or_peptide / ID: 51 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 17.743707 KDa |
| Sequence | String: MGIQQITNMC SHLQNASRAR LGLTSLPNTK YNLALALALH RAGFISSITR GGPHPPTPEA LMTYEPEPVT SANVATRRLW VGLKYWNEE PVLKELKPIS KPSRLVTASL EQLNRVARGF PAGYMKGLQL GECLFVNTDR GVLEVREAVE RKVGGLVLCK V K UniProtKB: 37S ribosomal protein S8, mitochondrial |
+Macromolecule #52: uS9m
| Macromolecule | Name: uS9m / type: protein_or_peptide / ID: 52 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 35.31957 KDa |
| Sequence | String: MMASLRHSIT SALRSSRQGC SKSAQWQSLD QQFGALRISS RSLSTHAHDH NPNVDGQFIA APALSIDNFK LHPYARAVPV SPSYFTRTP RFYDNYLSLE KLMEEYEDLP VIPATAVERV AWKTLEDIRK ELGEQVKASE FARCLALVKR LNSIHPDLKP Q EIKDVLDS ...String: MMASLRHSIT SALRSSRQGC SKSAQWQSLD QQFGALRISS RSLSTHAHDH NPNVDGQFIA APALSIDNFK LHPYARAVPV SPSYFTRTP RFYDNYLSLE KLMEEYEDLP VIPATAVERV AWKTLEDIRK ELGEQVKASE FARCLALVKR LNSIHPDLKP Q EIKDVLDS FRRNVQPFTN VAKPIPIDQF GRACGVGKRK ASSARAFVVE GTGEVLVNGK TLAEYFGRVH DRESAVWALR AT NRIDKYN VWVKVEGGGT TGQAEAITLA IAKALLAHEP ALKPALRRAG CVTRDPRKVE RKKHGHVKAR KMPTWVKR UniProtKB: 37S ribosomal protein S9, mitochondrial |
+Macromolecule #53: 30S ribosomal protein S10, mitochondrial
| Macromolecule | Name: 30S ribosomal protein S10, mitochondrial / type: protein_or_peptide / ID: 53 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 30.37707 KDa |
| Sequence | String: MIIRPVVRSL QGRAPLATLR SAQSVFQPLP QLRRNASVTT GPDAASKAST TPIARITTTT EAPKDQKKAK AAAAAAAEPE EQEVIMSRM PRSLEALYLQ PLRREAEYGV PSCDLQLRSY SIRNLEFFCD FALRAAYYLG LPAFGPVPLP RMIERWTVPK S HFIFKKSQ ...String: MIIRPVVRSL QGRAPLATLR SAQSVFQPLP QLRRNASVTT GPDAASKAST TPIARITTTT EAPKDQKKAK AAAAAAAEPE EQEVIMSRM PRSLEALYLQ PLRREAEYGV PSCDLQLRSY SIRNLEFFCD FALRAAYYLG LPAFGPVPLP RMIERWTVPK S HFIFKKSQ ENFERVTLRR LIQIKDGHPE TVQLWLAFLQ KHAYYGIGMK ANVWEFSKLG VSKAMDESKS EVEKLLETRW EH LSHVSDM KGVGNFEDFL AKERLRISGG R UniProtKB: 30S ribosomal protein S10, mitochondrial |
+Macromolecule #54: Translational machinery component
| Macromolecule | Name: Translational machinery component / type: protein_or_peptide / ID: 54 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 42.159832 KDa |
| Sequence | String: MAGQIDPIRA ELVGWTPRVP RNLLASLPPL GRKTAPKLPQ NISFSPPLSA HPCYQCQEKC LPHRRPVSWS PLILQRCRLR CLPRGHPLK ISRTFELPVG ERDGSHPLTA FARAPRTTEP RSLHYLYDSL PPTPALCAQL LFHQLIQGHF SFVVHPTVAT A PRSIWPAK ...String: MAGQIDPIRA ELVGWTPRVP RNLLASLPPL GRKTAPKLPQ NISFSPPLSA HPCYQCQEKC LPHRRPVSWS PLILQRCRLR CLPRGHPLK ISRTFELPVG ERDGSHPLTA FARAPRTTEP RSLHYLYDSL PPTPALCAQL LFHQLIQGHF SFVVHPTVAT A PRSIWPAK MSRFSTGRLL AQNLFQAFNK PVFPSATPVW TRAFSQTAAR RDADSESSAK LMESLTRGIV GMAADPTDLT GD KLATNIG LRDTEDEPYH FHIYSHKHNT HITVTKPNRD ALISLSCGNL GFKKSNRKHY DSAYQLGAYV VDKMHQMNLH NKI KKMEVV LRGFGPGREA VIKVLLGNEG RMLRSSIVRV SDATRLKFGG TRSKKPRRLG UniProtKB: Translational machinery component |
+Macromolecule #55: Ribosomal protein S12
| Macromolecule | Name: Ribosomal protein S12 / type: protein_or_peptide / ID: 55 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 18.657877 KDa |
| Sequence | String: MATTFLRNLF SQAQPSLLRR PLMPSSSSIL PAAARLFSST PAQNATLNQV MRGCRKPQRA RHAVSPALSS IKSPALKGVC VKVGITRPK KPNSGERKTA RVRLSTGKVI TAYIPGEGHN ISQHSVVLVR GGRAQDCPGV RYHLVRGALD LAGVATRMSS R SKYGTKKP KKASVG UniProtKB: Ribosomal protein S12 |
+Macromolecule #56: Probable ribosomal protein S13
| Macromolecule | Name: Probable ribosomal protein S13 / type: protein_or_peptide / ID: 56 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 13.503821 KDa |
| Sequence | String: MVFILGVNFN EHKLVQKALE SFYGLGQQAS ARILAKYSIH PRAKMGTLPP KIVTALTAEL STMTIENDAR RLVLDNIKRL RDMGTYRGR RHAMGLPVRG QQTRNQIANA RKLNKIERHG UniProtKB: Small ribosomal subunit protein uS13m |
+Macromolecule #57: Mitochondrial 37S ribosomal protein MRP2
| Macromolecule | Name: Mitochondrial 37S ribosomal protein MRP2 / type: protein_or_peptide / ID: 57 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 13.210644 KDa |
| Sequence | String: MSMFRAKKLD LGCFTNVRVL RDHSKRKAFL EAEPERQALR YVIRNTTLPA RTRAVAQLQL TQMHAYTRPT QIRNRCILGG KSRGILRDF KMTRYNFRMN ALMGNIPGVK KASW UniProtKB: Mitochondrial 37S ribosomal protein MRP2 |
+Macromolecule #58: Related to ribosomal protein S15 (Mitochondrial)
| Macromolecule | Name: Related to ribosomal protein S15 (Mitochondrial) / type: protein_or_peptide / ID: 58 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 35.803988 KDa |
| Sequence | String: MPPRLPGPQG LRSLTLCLRP AVASPAQALQ PLIQTANISQ KEKKRKMKQD PYGWAQAQQR KAVNVKRQAE LQAQRDAAWG DPVKGITTP FVESFDSAGQ ASVSPPKVGP DGQLVEEPKP LPTSPHLRNY LLNKDEFDSA IQYAEHILKP IKAEDRLTAD P EKEDEEAR ...String: MPPRLPGPQG LRSLTLCLRP AVASPAQALQ PLIQTANISQ KEKKRKMKQD PYGWAQAQQR KAVNVKRQAE LQAQRDAAWG DPVKGITTP FVESFDSAGQ ASVSPPKVGP DGQLVEEPKP LPTSPHLRNY LLNKDEFDSA IQYAEHILKP IKAEDRLTAD P EKEDEEAR EHAARHAKAV AALERIAKLE HGGAKDRKHA NIRRCIETFG RHITDQSLER PTPPLARGVE PKPQPVRAGP DT GSSEVQI AILTSKIRAL SKALEGHGGN RDKNNKRSLR RLCHKRQRLL RYMERKERGS GRWHHMLETL GLTPATWKGQ ITL UniProtKB: Related to ribosomal protein S15 (Mitochondrial) |
+Macromolecule #59: bS16m
| Macromolecule | Name: bS16m / type: protein_or_peptide / ID: 59 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 12.140166 KDa |
| Sequence | String: MVLKLRLARF GRTNAPFYNI VVAHARTARN SKPLEVIGTY DPVPKKDTYD TTGKLHKDIK LDVTRAKYWI GVGAQPTDTV WRLLSLVGI LDPKYKKPQP AQEQKTKA UniProtKB: Ribosomal protein S16 |
+Macromolecule #60: uS17m
| Macromolecule | Name: uS17m / type: protein_or_peptide / ID: 60 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 18.505701 KDa |
| Sequence | String: MASTIAAATK VSRKMFRELH GVVVTSGLID KTVKVRVGGQ KFNKFLQKHF DDPKQYLVHD PNNSLRAGDV VAIMPGFITS KSKRHVVKH IIAPAGTPIE ERPPIPSLDE LWDAKDAAKA AKKERKVLRE KMQAAEEAIE LAERMARHAV REIAMREKII S LQKVD UniProtKB: Nucleic acid-binding protein |
+Macromolecule #61: Ribosomal protein S18
| Macromolecule | Name: Ribosomal protein S18 / type: protein_or_peptide / ID: 61 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 28.468594 KDa |
| Sequence | String: MSSSRQFLSS ALRQCRAAGQ RTAQFSTTSP AAAIRDIPTT SFQNAPRTNT NTSSPSSNNN NNAGSSQYKS DMFNLLNSGA GSRGGRDSY NNARDVDGGQ RYGSNSQKVQ ELLQSEVTSN NYMRMMRRRW NNGDVYAPRD LSPYEMKGRN GGWVEAADVV D ELGFSPLD ...String: MSSSRQFLSS ALRQCRAAGQ RTAQFSTTSP AAAIRDIPTT SFQNAPRTNT NTSSPSSNNN NNAGSSQYKS DMFNLLNSGA GSRGGRDSY NNARDVDGGQ RYGSNSQKVQ ELLQSEVTSN NYMRMMRRRW NNGDVYAPRD LSPYEMKGRN GGWVEAADVV D ELGFSPLD NYRNFSLISD YITPFGRIRH SKETGLKPVN QRKIAKMVRR AIGLGIHPSV HKHPEILKNS NGRHLLPLVV PK GQKANKT KNFDGEDEN UniProtKB: Small ribosomal subunit protein bS18m |
+Macromolecule #62: Ribosomal protein S19/S15
| Macromolecule | Name: Ribosomal protein S19/S15 / type: protein_or_peptide / ID: 62 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 10.408332 KDa |
| Sequence | String: MQPTRVLFKR SVWKGPHIVP LPIQRPEPGK KIAPIRTQAR SATILPNFVG LKFQVHNGKD YIDLTVTEEM VGHKLGEFSA TRKPFIWGK KK UniProtKB: Ribosomal protein S19/S15 |
+Macromolecule #63: bS21m
| Macromolecule | Name: bS21m / type: protein_or_peptide / ID: 63 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 25.679785 KDa |
| Sequence | String: MELRSALQSV CRTTAAAASA SARSSLAGRH AFSTSARFQL QPPPNPYVSG KSRIETLRAA LSNRKPAAGA AAGGSPGATS ASSALPPRP PTANDSPWSI VDAINKDSNS STSSSSSSSS SGGALYSSSK PSPMQTWNET DFETRHMAPQ QELNIRLRPS T GRTFYVSG ...String: MELRSALQSV CRTTAAAASA SARSSLAGRH AFSTSARFQL QPPPNPYVSG KSRIETLRAA LSNRKPAAGA AAGGSPGATS ASSALPPRP PTANDSPWSI VDAINKDSNS STSSSSSSSS SGGALYSSSK PSPMQTWNET DFETRHMAPQ QELNIRLRPS T GRTFYVSG HQDFAGALKL LHRTVAANKV KKDVRLQRFH ERPALKRKRN LRERWRARFK EGFKAAVNRT FELKNQGW UniProtKB: Ribosomal protein S21 |
+Macromolecule #64: 37S ribosomal protein S25, mitochondrial
| Macromolecule | Name: 37S ribosomal protein S25, mitochondrial / type: protein_or_peptide / ID: 64 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 29.553588 KDa |
| Sequence | String: MGKGRIFQAA KVYQRASEAA ATNIVSGLPP RNPPLWLKAI ESIPPAEINT RPYPIQHSPP NPRARKPRNL FRPTKIVYPE DELRRDFYR DHPWELARPR MIIELDGKDA RFLDWSKGLR QRGIPLSGER HGNWMLTRIA NDSVVQRQLW LMENQGMTKQ E AYDKARHE ...String: MGKGRIFQAA KVYQRASEAA ATNIVSGLPP RNPPLWLKAI ESIPPAEINT RPYPIQHSPP NPRARKPRNL FRPTKIVYPE DELRRDFYR DHPWELARPR MIIELDGKDA RFLDWSKGLR QRGIPLSGER HGNWMLTRIA NDSVVQRQLW LMENQGMTKQ E AYDKARHE FYKLRQLEQM ERRIAVEEAR MVGGYFGKDL LTVGMELENK TYESWKKWAT TEIARQESAR ASMYTNVVDN SA LEESEED ELLAQN UniProtKB: Small ribosomal subunit protein mS23 |
+Macromolecule #65: mS26
| Macromolecule | Name: mS26 / type: protein_or_peptide / ID: 65 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 35.312184 KDa |
| Sequence | String: MAPTLARPSL SGVQFILSSP TTTCAATSVV TRAIAARSFS TTRSARDSVS IPPDSPNYIK VPEPPQSSEV RHPFVKGHLP IPRSIFPKK GVPEKVQSGY VNRIAPKSAA ELAGLPPKSK QESWRRKMAE ARRQSLEAGL QGLWQRKVKR DQKQAKESKA R YLANKRAA ...String: MAPTLARPSL SGVQFILSSP TTTCAATSVV TRAIAARSFS TTRSARDSVS IPPDSPNYIK VPEPPQSSEV RHPFVKGHLP IPRSIFPKK GVPEKVQSGY VNRIAPKSAA ELAGLPPKSK QESWRRKMAE ARRQSLEAGL QGLWQRKVKR DQKQAKESKA R YLANKRAA QAPERLDEVF TRATIRESTA KNTFVPLDPE AFVKAEEARI KHAEKEAMKS EARRDAVVQL YVASKNFIVD EK ELEEHVN KHFTEKIHNA GLWESGRSIW DSQKNPISMR ELRNEFSGFN DRVTATTSAA VKTTVRQKNV AEELTGGKL UniProtKB: Small ribosomal subunit protein mS26 |
+Macromolecule #66: mS27
| Macromolecule | Name: mS27 / type: protein_or_peptide / ID: 66 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 44.923723 KDa |
| Sequence | String: MAGRAPQHAL RVGCRAVPEA LSKPAQQSRC LSSTVPRQAT YPVVSFNKTS SPELKEALET LREKVILPTY LPPELRQKIF NKKYEKELA HDPVTIQIDG QPQRFSYINM LTDMPNTPKN IRAALLSMKN GGDFANLSGL LEGMHRANRK LPYWLSAQIV R KACKAGHL ...String: MAGRAPQHAL RVGCRAVPEA LSKPAQQSRC LSSTVPRQAT YPVVSFNKTS SPELKEALET LREKVILPTY LPPELRQKIF NKKYEKELA HDPVTIQIDG QPQRFSYINM LTDMPNTPKN IRAALLSMKN GGDFANLSGL LEGMHRANRK LPYWLSAQIV R KACKAGHL QLILNMVRDV KRTGFTLERH ETVNELLFWI QRFAWKSDYS EPETRKALRE VQEILDALEG DERHMSKDRK RQ QALTRFP YHRDPQFLAA RLNLTAELAA RRAVTGQTSE QQLNSANDVK NLVKYAEQLV RLWPADKALL DMYTDEAYVA RVD LRYLIK PQVHLRYASF TLQALKNAAK IVGQLGHGPL AAQLINRAAA VEAESQLAYA KVDDGMAGQK IYEMVVGGKK UniProtKB: Small ribosomal subunit protein mS27 |
+Macromolecule #67: Mitochondrial ribosomal protein DAP3
| Macromolecule | Name: Mitochondrial ribosomal protein DAP3 / type: protein_or_peptide / ID: 67 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 51.451414 KDa |
| Sequence | String: MSASNCLRCL VRPSAVALVP RQMLQVGGGS LTFTATAATK GASSANTGGR QNANRPQKLR FKKTKKKNVV SSGKPPLPGE RKAYRKKIV LSNNNALPVP GLETLRPNDL AKQDNVGSVK ALPEDVVDAL RAMEAFKPTQ CWGIFRQPSV LIRQETVDLT K KMKAAGAD ...String: MSASNCLRCL VRPSAVALVP RQMLQVGGGS LTFTATAATK GASSANTGGR QNANRPQKLR FKKTKKKNVV SSGKPPLPGE RKAYRKKIV LSNNNALPVP GLETLRPNDL AKQDNVGSVK ALPEDVVDAL RAMEAFKPTQ CWGIFRQPSV LIRQETVDLT K KMKAAGAD GKTIRMVIEG NRVTGKSLLL LQAMTHAFMN DWVVLHIPEA QELTTAVTEY APIENSPLWT QPTYTLKLLQ SF KRANEKV LSRMNTVYSH ADLPQIIPVN SPLLQLINSA KEADGAWTVF QALWRELNAE NVPGRPPILF SLDGLAHIMK VSD YRNPAF ELIHSHDLAL VKLFTDCLSG ATVMPNGGAV LGATTRGNSP RSASMELAIA QREAEKAGEK EVPQRDPYSK KYDD RVEAV MKSVEILRLK GVSKTEARGL LEYWAASGML KKRVDESMVS EKWTLSGNGV VGEMERASLL TMKA UniProtKB: Small ribosomal subunit protein mS29 |
+Macromolecule #68: mS33
| Macromolecule | Name: mS33 / type: protein_or_peptide / ID: 68 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 12.169154 KDa |
| Sequence | String: MSVPRARLLQ LVKARCELFS TTFNPEGIRT GNKILRQRLK GPALATYYPR KNVGIRELQK EFGTLGLEVD DEVDDDRLEH LAALRARDK GAPKKKRTAP SAADAKKKK UniProtKB: Mitochondrial 37S ribosomal protein S27 |
+Macromolecule #69: 37S ribosomal protein S24, mitochondrial
| Macromolecule | Name: 37S ribosomal protein S24, mitochondrial / type: protein_or_peptide / ID: 69 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 44.05709 KDa |
| Sequence | String: MASSANALRL GLRTCSRPTQ TSNLRIAALQ RRALSATATR CEGVPVPKRE DDAKLEFTRP YNPGEFLSEK LRSDDLQDWE RERYERALT SWEQTPDDLK RGWSTMIRDI EQAAAPLRRV VMPRRSTFWY EEEKDTDLIT NEDGEDDFHE NDIMSLGHGK L EEHREFRE ...String: MASSANALRL GLRTCSRPTQ TSNLRIAALQ RRALSATATR CEGVPVPKRE DDAKLEFTRP YNPGEFLSEK LRSDDLQDWE RERYERALT SWEQTPDDLK RGWSTMIRDI EQAAAPLRRV VMPRRSTFWY EEEKDTDLIT NEDGEDDFHE NDIMSLGHGK L EEHREFRE YARIAVWEMP LLSKYAKPFV PPTSEEVLRF RYTTYMGEFH PADRKVVVEF CPKDLRDLSE VQQRKLMKLA GP RYNPEKD IIKMSCEKFE HQAQNKRYLG DLIEKMIAAA KDPKDTFEDI PLDTRHHTFT KKISFPKEWL LTEERKKELE AAR QQALLK DAEKVVQGAL VDGADVVKQY LESGAAEALH AVPVMAGRGG KALPGGKGGK MQRSKR UniProtKB: Small ribosomal subunit protein mS35 |
+Macromolecule #70: 37S ribosomal protein mrp10, mitochondrial
| Macromolecule | Name: 37S ribosomal protein mrp10, mitochondrial / type: protein_or_peptide / ID: 70 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 9.907534 KDa |
| Sequence | String: MPNKPIRLPP LKQLRVRQAN KAEENPCIAV MSSVLACWAS AGYNSAGCAT VENALRACMD APKPAPKPNN TINYHLSRFQ ERLTQGKSK K UniProtKB: 37S ribosomal protein mrp10, mitochondrial |
+Macromolecule #71: DUF1713 domain-containing protein
| Macromolecule | Name: DUF1713 domain-containing protein / type: protein_or_peptide / ID: 71 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 38.12625 KDa |
| Sequence | String: MIPQSVRRVV AAAPQSPVVS SLAASSAPRA GASYILSSYQ PNALQKRRYS SSKPSSPDDG SSRAFAARAS VPAAGTSKTP GEKRKRKAK EPVMPPLPSV PSTRHIKDEA LALSTFFALH RPISVTQLLP KTVTEESFAE IFNHRKGHKA TDVLSTLSQA V HDLESPMS ...String: MIPQSVRRVV AAAPQSPVVS SLAASSAPRA GASYILSSYQ PNALQKRRYS SSKPSSPDDG SSRAFAARAS VPAAGTSKTP GEKRKRKAK EPVMPPLPSV PSTRHIKDEA LALSTFFALH RPISVTQLLP KTVTEESFAE IFNHRKGHKA TDVLSTLSQA V HDLESPMS GLRLSDNDVE DGSTKISLKH PDGTESDVYF QLNSMSGHFL PFNPPPPPEP IAEVDEAAAE AEASAAIAEE IA ATAEQEP ETRVYKAVVT IEETRDTNGQ YKIMAHSPQL VEDDAVQPRS FLERLALRQI RYEEARQQQG IMHAISVRRQ KKL KIKKKK YKKLMRRTRN ERRKQDRL UniProtKB: Small ribosomal subunit protein mS38 |
+Macromolecule #72: Protein FYV4, mitochondrial
| Macromolecule | Name: Protein FYV4, mitochondrial / type: protein_or_peptide / ID: 72 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 26.612898 KDa |
| Sequence | String: MKSSLRIQST LGLLFRPCAV SAPATAPGLN AHARWAHKTA AQLPLIPKPT PFVPDVPTFL TLIGRDLKQH ADKFPTWEAL FTLTTDQLR ELGVEPPRAR RYLLRWRQRF REGKFGIGGD LKHVENGVAY LKIHEKEASP TRTSRRVVNV PANQHVEEVS E GERVKVKG ...String: MKSSLRIQST LGLLFRPCAV SAPATAPGLN AHARWAHKTA AQLPLIPKPT PFVPDVPTFL TLIGRDLKQH ADKFPTWEAL FTLTTDQLR ELGVEPPRAR RYLLRWRQRF REGKFGIGGD LKHVENGVAY LKIHEKEASP TRTSRRVVNV PANQHVEEVS E GERVKVKG YKVKGVSTIV GPYALPVQKG VAKLAVTEGM WEDKRGHKVD GGERRRAEVR FKRGVAERKA LREKMGFY UniProtKB: Small ribosomal subunit protein mS41 |
+Macromolecule #73: Manganese and iron superoxide dismutase
| Macromolecule | Name: Manganese and iron superoxide dismutase / type: protein_or_peptide / ID: 73 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 34.270785 KDa |
| Sequence | String: MIGSMGLRIP RATSSLLGRA VNAVATNQSA VAGASIAAQR RWQHYLMPLR DNFEQEGIRN FLSPGSVNMA YTEYQTFILE KLNALVVGT DFEQKDTKSI VLATARDPEL AHVFNHASMA HNNHFFFDHL SPVPVKMGDK LFYHINENFG SVDTLRDEMI G TAVSMFGP ...String: MIGSMGLRIP RATSSLLGRA VNAVATNQSA VAGASIAAQR RWQHYLMPLR DNFEQEGIRN FLSPGSVNMA YTEYQTFILE KLNALVVGT DFEQKDTKSI VLATARDPEL AHVFNHASMA HNNHFFFDHL SPVPVKMGDK LFYHINENFG SVDTLRDEMI G TAVSMFGP GFVWLVRTQL PGQPVALRVM ATYLAGSPYP GAHWRRQEMD AQTSIGSSPQ GLSNGQRFFE RSAAGFKGNK LE PTAPGGT DLIPILCLNT WEYAWLREYG TGVGGMGGKL AYAQSWWNMI DWAKVEEEAR LETRILTGGD SSV UniProtKB: Manganese and iron superoxide dismutase |
+Macromolecule #74: mS45
| Macromolecule | Name: mS45 / type: protein_or_peptide / ID: 74 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 39.274625 KDa |
| Sequence | String: MPPRIPNAPS SLLLQSSACS GSSSRCTASR LPTWASSCSS SSPQSSQPTT HQQQCSSFST TAPAHVQSVR RQKMFSWLDK KGSAYKEHT RQGPNLLGGQ GKDGLAVPFP NNPYFKSQPV LSEGSREIIY QDVMEKGLPI KAVSAKYNVD VRRVAAVIRL K EIEKRWIK ...String: MPPRIPNAPS SLLLQSSACS GSSSRCTASR LPTWASSCSS SSPQSSQPTT HQQQCSSFST TAPAHVQSVR RQKMFSWLDK KGSAYKEHT RQGPNLLGGQ GKDGLAVPFP NNPYFKSQPV LSEGSREIIY QDVMEKGLPI KAVSAKYNVD VRRVAAVIRL K EIEKRWIK EYKPLARPYA RAVMKMLPQT VLGGPDQKPH ESINDVHVHS YTTQQLFVPV SESREFTRED AAKAFGDHIL PV DKKLRVP ELIEFQKDLL KEVPLQEANR KFLNATAASE AKIAEREAKR RQAVEDAITR VKTDRFEFRF QEFNAENVGH DGR DRNAVG WRYGVPFPDR KRSQIKIPTK VE UniProtKB: Small ribosomal subunit protein mS45 |
+Macromolecule #75: mS46
| Macromolecule | Name: mS46 / type: protein_or_peptide / ID: 75 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 43.112672 KDa |
| Sequence | String: MNRQVVTSTL GRRGVASTIL NAQQQQRPFS STTTRCAAED DSKKPAAAPS TPRAAAPGPI SASRQKSEAA VGKLTQLRGS FTSLTNDNS FHKTLPAGAR DARRLAAAPI AGKGAGAGAV APLGGGGGAS GAPKVINVRS LKGTLGSRGS NNIPGAVAPG A ALRPRFAA ...String: MNRQVVTSTL GRRGVASTIL NAQQQQRPFS STTTRCAAED DSKKPAAAPS TPRAAAPGPI SASRQKSEAA VGKLTQLRGS FTSLTNDNS FHKTLPAGAR DARRLAAAPI AGKGAGAGAV APLGGGGGAS GAPKVINVRS LKGTLGSRGS NNIPGAVAPG A ALRPRFAA GPGAAAGRPR FGAAASPGAG PTGAARRPPF GARRARPAGD KKRSGGSGDK RPRGDDYDAP PTEEEKAFLR GL EQGKVTE YVPKLTPDTL LGYGPPVATD AALGKVESAM RTMRILGGGL PFNDQSGVTS DPTAIKHRYV HEKKPVFFSS VEE KEWVRE SLDKFAVSEG PEKKTKQKIL ETSVLGKYEE PKYVESLTET VKMVEKYQGG TFSYAPSDAD KFNKKLNQLL AAGL PRAAP APAQAQKKA UniProtKB: Protein FYV4, mitochondrial |
+Macromolecule #76: 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial
| Macromolecule | Name: 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial / type: protein_or_peptide / ID: 76 / Number of copies: 1 / Enantiomer: LEVO / EC number: 3-hydroxyisobutyryl-CoA hydrolase |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 56.401191 KDa |
| Sequence | String: MLVSKSIASR AGLASAVASR SCRPSFTAMP LRAKVLGQQA PISTQAAAEL FKPVEGDEPE DVLFNSLYNL RSVELNRPAK YNALNGSMI RKIAPRLLEW ERSDMANVIV IKGSGEKAFC AGGDVAALAK QNAEGPEGVK KSVDYFGLEY KLNHLISTYT R PYVAFLDG ...String: MLVSKSIASR AGLASAVASR SCRPSFTAMP LRAKVLGQQA PISTQAAAEL FKPVEGDEPE DVLFNSLYNL RSVELNRPAK YNALNGSMI RKIAPRLLEW ERSDMANVIV IKGSGEKAFC AGGDVAALAK QNAEGPEGVK KSVDYFGLEY KLNHLISTYT R PYVAFLDG ITMGGGVGLS IHAPFRIATE RTVFAMPETK IGFFPDVGAS FFLPRMPGQV GPYLGLTSAL LKGVQVYYAG IA THYLHSS SLPALESRLA ELTPRDYWTI EQRLSVINDT IEEFSTGVPY DENIEIGGKI RLAIDRCFKY DKIDEIIAAL KEE AAEGAK GGVQSWAKNT LEELTQRSPT SLHVTLRQMR LGKSWGIAHT FKREHQMAAK FMKSHDFNEG VTALLIDKGA NGPA KWKPA SLDEIPPGAN ISEDYFRNDP EVPVLELLND RSYMQYPYNK FGLPNDYDVK EAIEKGNFTR EKLIDHFVET RRGKQ GVRE AVSDVLDRMA VRSKGTEHVQ WKKE UniProtKB: 3-hydroxyisobutyryl-CoA hydrolase |
+Macromolecule #77: IF1
| Macromolecule | Name: IF1 / type: protein_or_peptide / ID: 77 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 10.664098 KDa |
| Sequence | String: MLRTTVSKLA RPTVSRAFAT TSRALAGETG APPKTGGPGD AFQRREKANE DFAIRQREKE KLLELKKKLA EQQKHLKTLS DHIDEITRE QGGERN UniProtKB: ATP synthase F1 subunit epsilon |
+Macromolecule #81: Poly-Peptide
| Macromolecule | Name: Poly-Peptide / type: protein_or_peptide / ID: 81 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 515.56 Da |
| Sequence | String: AAAAAAA |
+Macromolecule #82: 60S ribosomal protein L1, mitochondrial
| Macromolecule | Name: 60S ribosomal protein L1, mitochondrial / type: protein_or_peptide / ID: 82 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 33.327664 KDa |
| Sequence | String: MASTQQCLAS LARLSLSTPT RAALPTIPKF LVPSVAASQV RYATNNPNKG GAKKAPKKKK QYKFFKSWDL TGQQQFSLCD AMRYLRAVE VGQPPLSVKY EVHVKLRTKK NGPVVRDRVR LPTPVKTDTR IAVICPEGSA LQEEAKNLGA VMAGEETLFE A IRSGNFPF ...String: MASTQQCLAS LARLSLSTPT RAALPTIPKF LVPSVAASQV RYATNNPNKG GAKKAPKKKK QYKFFKSWDL TGQQQFSLCD AMRYLRAVE VGQPPLSVKY EVHVKLRTKK NGPVVRDRVR LPTPVKTDTR IAVICPEGSA LQEEAKNLGA VMAGEETLFE A IRSGNFPF NKLLCHTESE GALRKANVGK LLGPKGLMPS GKTKTITNNL EATFRDMIGM DEYRERNGVV RMAVGQLGFT PK QLAENIR VFMAKIKSDI GKLDDTTPKM VEEVVLSTTH GPGMSLNAEF APTDDKIKPE DLESVM UniProtKB: Related to ribosomal protein L1 |
+Macromolecule #83: L51_S25_CI-B8 domain-containing protein
| Macromolecule | Name: L51_S25_CI-B8 domain-containing protein / type: protein_or_peptide / ID: 83 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 22.734252 KDa |
| Sequence | String: MVGVIRRLNV VRELLNIRAG PGAALLPKEV TKVHMQFAHR IEEGHMGPRK FWRENLPKLK YWNPAVPMVI NRTTDQKGPA VMTIYFRDD KDAKPSSTPF PTSSADGSSP APKPAQGERI VTIDMKNRNS SVILKEFLDK TGAVAVQPTA KDEEEFREFE D LHRRSEID ...String: MVGVIRRLNV VRELLNIRAG PGAALLPKEV TKVHMQFAHR IEEGHMGPRK FWRENLPKLK YWNPAVPMVI NRTTDQKGPA VMTIYFRDD KDAKPSSTPF PTSSADGSSP APKPAQGERI VTIDMKNRNS SVILKEFLDK TGAVAVQPTA KDEEEFREFE D LHRRSEID RERIRKMNDA KKREKAMLAK AMSDAQSIKA ASA UniProtKB: Large ribosomal subunit protein mL61 |
+Macromolecule #84: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 84 / Number of copies: 250 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
+Macromolecule #85: SPERMINE
| Macromolecule | Name: SPERMINE / type: ligand / ID: 85 / Number of copies: 1 / Formula: SPM |
|---|---|
| Molecular weight | Theoretical: 202.34 Da |
| Chemical component information |  ChemComp-SPM: |
+Macromolecule #86: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 86 / Number of copies: 49 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
+Macromolecule #87: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 87 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
+Macromolecule #88: NICOTINAMIDE-ADENINE-DINUCLEOTIDE
| Macromolecule | Name: NICOTINAMIDE-ADENINE-DINUCLEOTIDE / type: ligand / ID: 88 / Number of copies: 1 / Formula: NAD |
|---|---|
| Molecular weight | Theoretical: 663.425 Da |
| Chemical component information |  ChemComp-NAD: |
+Macromolecule #89: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 89 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2.7 | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-20 / Number real images: 3172 / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 130000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
+ About Yorodumi
About Yorodumi
-News
-Feb 9, 2022. New format data for meta-information of EMDB entries
New format data for meta-information of EMDB entries
- Version 3 of the EMDB header file is now the official format.
- The previous official version 1.9 will be removed from the archive.
Related info.: EMDB header
EMDB header
External links: wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
-Aug 12, 2020. Covid-19 info
Covid-19 info

URL: https://pdbj.org/emnavi/covid19.php
New page: Covid-19 featured information page in EM Navigator.
Related info.: Covid-19 info /
Covid-19 info /  Mar 5, 2020. Novel coronavirus structure data
Mar 5, 2020. Novel coronavirus structure data
+Mar 5, 2020. Novel coronavirus structure data
Novel coronavirus structure data
- International Committee on Taxonomy of Viruses (ICTV) defined the short name of the 2019 coronavirus as "SARS-CoV-2".
- In the structure databanks used in Yorodumi, some data are registered as the other names, "COVID-19 virus" and "2019-nCoV". Here are the details of the virus and the list of structure data.
Related info.: Yorodumi Speices /
Yorodumi Speices /  Aug 12, 2020. Covid-19 info
Aug 12, 2020. Covid-19 info
External links: COVID-19 featured content - PDBj /
COVID-19 featured content - PDBj /  Molecule of the Month (242):Coronavirus Proteases
Molecule of the Month (242):Coronavirus Proteases
+Jan 31, 2019. EMDB accession codes are about to change! (news from PDBe EMDB page)
EMDB accession codes are about to change! (news from PDBe EMDB page)
- The allocation of 4 digits for EMDB accession codes will soon come to an end. Whilst these codes will remain in use, new EMDB accession codes will include an additional digit and will expand incrementally as the available range of codes is exhausted. The current 4-digit format prefixed with “EMD-” (i.e. EMD-XXXX) will advance to a 5-digit format (i.e. EMD-XXXXX), and so on. It is currently estimated that the 4-digit codes will be depleted around Spring 2019, at which point the 5-digit format will come into force.
- The EM Navigator/Yorodumi systems omit the EMD- prefix.
Related info.: Q: What is EMD? /
Q: What is EMD? /  ID/Accession-code notation in Yorodumi/EM Navigator
ID/Accession-code notation in Yorodumi/EM Navigator
External links: EMDB Accession Codes are Changing Soon! /
EMDB Accession Codes are Changing Soon! /  Contact to PDBj
Contact to PDBj
+Jul 12, 2017. Major update of PDB
Major update of PDB
- wwPDB released updated PDB data conforming to the new PDBx/mmCIF dictionary.
- This is a major update changing the version number from 4 to 5, and with Remediation, in which all the entries are updated.
- In this update, many items about electron microscopy experimental information are reorganized (e.g. em_software).
- Now, EM Navigator and Yorodumi are based on the updated data.
External links: wwPDB Remediation /
wwPDB Remediation /  Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
-Yorodumi
Thousand views of thousand structures
- Yorodumi is a browser for structure data from EMDB, PDB, SASBDB, etc.
- This page is also the successor to EM Navigator detail page, and also detail information page/front-end page for Omokage search.
- The word "yorodu" (or yorozu) is an old Japanese word meaning "ten thousand". "mi" (miru) is to see.
Related info.: EMDB /
EMDB /  PDB /
PDB /  SASBDB /
SASBDB /  Comparison of 3 databanks /
Comparison of 3 databanks /  Yorodumi Search /
Yorodumi Search /  Aug 31, 2016. New EM Navigator & Yorodumi /
Aug 31, 2016. New EM Navigator & Yorodumi /  Yorodumi Papers /
Yorodumi Papers /  Jmol/JSmol /
Jmol/JSmol /  Function and homology information /
Function and homology information /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
 Movie
Movie Controller
Controller

















































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)