[English] 日本語
 Yorodumi
Yorodumi- EMDB-10845: Cryo-EM structure of yeast mitochondrial RNA polymerase transcrip... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10845 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription pre-initiation complex | |||||||||
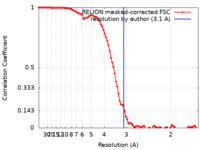
 Map data Map data | post process map 3.1A | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | gene transcription / polymerase / RDRP / MTF1 / RPO41 / POLRMT / mtRNAP / DNA / transcription initiation / RNA polymerase / mitochondria / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationMitochondrial transcription initiation / mitochondrial DNA-directed RNA polymerase complex / mitochondrial transcription factor activity / mitochondrial promoter sequence-specific DNA binding / transcription initiation at mitochondrial promoter / : / mitochondrial transcription / DNA replication, synthesis of primer / positive regulation of DNA-templated transcription, elongation / mitochondrial nucleoid ...Mitochondrial transcription initiation / mitochondrial DNA-directed RNA polymerase complex / mitochondrial transcription factor activity / mitochondrial promoter sequence-specific DNA binding / transcription initiation at mitochondrial promoter / : / mitochondrial transcription / DNA replication, synthesis of primer / positive regulation of DNA-templated transcription, elongation / mitochondrial nucleoid / Transferases; Transferring one-carbon groups; Methyltransferases / methyltransferase activity / mitochondrial intermembrane space / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / methylation / mitochondrial matrix / mitochondrion / DNA binding / RNA binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Das K | |||||||||
| Funding support |  Belgium, 1 items Belgium, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2021 Journal: Mol Cell / Year: 2021Title: Cryo-EM Structures Reveal Transcription Initiation Steps by Yeast Mitochondrial RNA Polymerase. Authors: Brent De Wijngaert / Shemaila Sultana / Anupam Singh / Chhaya Dharia / Hans Vanbuel / Jiayu Shen / Daniel Vasilchuk / Sergio E Martinez / Eaazhisai Kandiah / Smita S Patel / Kalyan Das /    Abstract: Mitochondrial RNA polymerase (mtRNAP) is crucial in cellular energy production, yet understanding of mitochondrial DNA transcription initiation lags that of bacterial and nuclear DNA transcription. ...Mitochondrial RNA polymerase (mtRNAP) is crucial in cellular energy production, yet understanding of mitochondrial DNA transcription initiation lags that of bacterial and nuclear DNA transcription. We report structures of two transcription initiation intermediate states of yeast mtRNAP that explain promoter melting, template alignment, DNA scrunching, abortive synthesis, and transition into elongation. In the partially melted initiation complex (PmIC), transcription factor MTF1 makes base-specific interactions with flipped non-template (NT) nucleotides "AAGT" at -4 to -1 positions of the DNA promoter. In the initiation complex (IC), the template in the expanded 7-mer bubble positions the RNA and NTP analog UTPαS, while NT scrunches into an NT loop. The scrunched NT loop is stabilized by the centrally positioned MTF1 C-tail. The IC and PmIC states coexist in solution, revealing a dynamic equilibrium between two functional states. Frequent scrunching/unscruching transitions and the imminent steric clashes of the inflating NT loop and growing RNA:DNA with the C-tail explain abortive synthesis and transition into elongation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10845.map.gz emd_10845.map.gz | 14.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10845-v30.xml emd-10845-v30.xml emd-10845.xml emd-10845.xml | 27.1 KB 27.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10845_fsc.xml emd_10845_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_10845.png emd_10845.png | 172.2 KB | ||
| Masks |  emd_10845_msk_1.map emd_10845_msk_1.map | 15.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10845.cif.gz emd-10845.cif.gz | 7.8 KB | ||
| Others |  emd_10845_half_map_1.map.gz emd_10845_half_map_1.map.gz emd_10845_half_map_2.map.gz emd_10845_half_map_2.map.gz | 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10845 http://ftp.pdbj.org/pub/emdb/structures/EMD-10845 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10845 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10845 | HTTPS FTP |
-Related structure data
| Related structure data |  6ymvMC  6ymwC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10845.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10845.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | post process map 3.1A | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.827 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10845_msk_1.map emd_10845_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-1
| File | emd_10845_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-2
| File | emd_10845_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : mitochondria DNA-dependent RNA polymerase pre-initiation complex
| Entire | Name: mitochondria DNA-dependent RNA polymerase pre-initiation complex |
|---|---|
| Components |
|
-Supramolecule #1: mitochondria DNA-dependent RNA polymerase pre-initiation complex
| Supramolecule | Name: mitochondria DNA-dependent RNA polymerase pre-initiation complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 202 KDa |
-Supramolecule #2: mitochondria DNA-dependent RNA polymerase pre-initiation complex
| Supramolecule | Name: mitochondria DNA-dependent RNA polymerase pre-initiation complex type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: DNA
| Supramolecule | Name: DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Mitochondrial transcription factor 1
| Macromolecule | Name: Mitochondrial transcription factor 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Transferases; Transferring one-carbon groups; Methyltransferases |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.151203 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGGSHHHHHH GMASSVPIPG IKDISKLKFF YGFKYLWNPT VYNKIFDKLD LTKTYKHPEE LKVLDLYPGV GIQSAIFYNK YCPRQYSLL EKRSSLYKFL NAKFEGSPLQ ILKRDPYDWS TYSNLIDEER IFVPEVQSSD HINDKFLTVA NVTGEGSEGL I MQWLSCIG ...String: MGGSHHHHHH GMASSVPIPG IKDISKLKFF YGFKYLWNPT VYNKIFDKLD LTKTYKHPEE LKVLDLYPGV GIQSAIFYNK YCPRQYSLL EKRSSLYKFL NAKFEGSPLQ ILKRDPYDWS TYSNLIDEER IFVPEVQSSD HINDKFLTVA NVTGEGSEGL I MQWLSCIG NKNWLYRFGK VKMLLWMPST TARKLLARPG MHSRSKCSVV REAFTDTKLI AISDANELKG FDSQCIEEWD PI LFSAAEI WPTKGKPIAL VEMDPIDFDF DVDNWDYVTR HLMILKRTPL NTVMDSLGHG GQQYFNSRIT DKDLLKKCPI DLT NDEFIY LTKLFMEWPF KPDILMDFVD MYQTEHSG UniProtKB: Mitochondrial transcription factor 1 |
-Macromolecule #2: DNA-directed RNA polymerase, mitochondrial
| Macromolecule | Name: DNA-directed RNA polymerase, mitochondrial / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 143.282656 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAMGSGIQRP SAVTSMTRTR DVMQLWSLLE ACLQSNLMKR AFSILESLYL VPEHKQRFIE DYNMYLNSFS KNDPNFPILK MNEKLTNDL ETSFKDVNYN DKTLAIMIHH ALNFHSTTSS MLLKPIISAY LKMSVNGIRE IFSCLDILTI SDLNILMNDL K VITPSQLP ...String: GAMGSGIQRP SAVTSMTRTR DVMQLWSLLE ACLQSNLMKR AFSILESLYL VPEHKQRFIE DYNMYLNSFS KNDPNFPILK MNEKLTNDL ETSFKDVNYN DKTLAIMIHH ALNFHSTTSS MLLKPIISAY LKMSVNGIRE IFSCLDILTI SDLNILMNDL K VITPSQLP NSVRPILESL TLSPTPVNNI ENEEGLNKVE AENDSKLHKA SNASSDSIKK PSLDPLREVS FHGSTEVLSK DA EKLIAVD TIGMRVIRHT LLGLSLTPEQ KEQISKFKFD ANDNVLKMKP TKNDDNNNSI NFFEIYNSLP TLEEKKAFES ALN IFNQDR QKVLENRATE AARERWKHDF EEAKARGDIS IEKNLNVKLW KWYNEMLPLV KEEINHCRSL LSEKLSDKKG LNKV DTNRL GYGPYLTLID PGKMCVITIL ELLKLNSTGG VIEGMRTARA VISVGKAIEM EFRSEQVLKS ESQAFRDVNK KSPEF KKLV QNAKSVFRSS QIEQSKILWP QSIRARIGSV LISMLIQVAK VSVQGVDPVT KAKVHGEAPA FAHGYQYHNG SKLGVL KIH KTLIRQLNGE RLIASVQPQL LPMLVEPKPW VNWRSGGYHY TQSTLLRTKD SPEQVAYLKA ASDNGDIDRV YDGLNVL GR TPWTVNRKVF DVVSQVWNKG EGFLDIPGAQ DEMVLPPAPP KNSDPSILRA WKLQVKTIAN KFSSDRSNRC DTNYKLEI A RAFLGEKLYF PHNLDFRGRA YPLSPHFNHL GNDMSRGLLI FWHGKKLGPS GLKWLKIHLS NLFGFDKLPL KDRVAFTES HLQDIKDSAE NPLTGDRWWT TADKPWQALA TCFELNEVMK MDNPEEFISH QPVHQDGTCN GLQHYAALGG DVEGATQVNL VPSDKPQDV YAHVARLVQK RLEIAAEKGD ENAKILKDKI TRKVVKQTVM TNVYGVTYVG ATFQIAKQLS PIFDDRKESL D FSKYLTKH VFSAIRELFH SAHLIQDWLG ESAKRISKSI RLDVDEKSFK NGNKPDFMSS VIWTTPLGLP IVQPYREESK KQ VETNLQT VFISDPFAVN PVNARRQKAG LPPNFIHSLD ASHMLLSAAE CGKQGLDFAS VHDSYWTHAS DIDTMNVVLR EQF IKLHEV DLVLRLKEEF DQRYKNYVKI GKLKRSTDLA QKIIRIRKDL SRKLGRSTTL ADEIYFEKKR QELLNSPLIE DRNV GEKMV TTVSLFEDIT DLDALELENG GDENSGMSVL LPLRLPEIPP KGDFDVTVLR NSQYFFS UniProtKB: DNA-directed RNA polymerase, mitochondrial |
-Macromolecule #3: DNA (33-MER) NON-TEMPLATE
| Macromolecule | Name: DNA (33-MER) NON-TEMPLATE / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.248671 KDa |
| Sequence | String: (DC)(DG)(DA)(DA)(DT)(DA)(DA)(DG)(DT)(DA) (DT)(DT)(DG)(DA)(DT)(DA)(DT)(DA)(DA)(DG) (DT)(DA)(DA)(DT)(DA)(DG)(DA)(DT)(DA) (DA)(DT)(DG)(DC) |
-Macromolecule #4: DNA (33-MER) template
| Macromolecule | Name: DNA (33-MER) template / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.047511 KDa |
| Sequence | String: (DG)(DC)(DA)(DT)(DT)(DA)(DT)(DC)(DT)(DA) (DC)(DC)(DG)(DA)(DC)(DA)(DA)(DT)(DA)(DT) (DC)(DA)(DA)(DT)(DA)(DC)(DT)(DT)(DA) (DT)(DT)(DC)(DG) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7 Details: 50mM Bis-tris propane, 100mM NaCl, 5mM MgCl2, 1mM EDTA, 2mM DTT |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 8 K / Instrument: LEICA EM GP / Details: 5 uL sample; back blotting for 12 -14 second. |
| Details | The sample was monodisperse with hydrodynamic radius 5.82nm. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 6.0 sec. / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 26.0 µm / Calibrated defocus min: 5.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 22.0 µm / Nominal defocus min: 7.0 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Details | Realspace refinement | ||||||||
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Coorelation coefficu=ient | ||||||||
| Output model |  PDB-6ymv: |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)