+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10746 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Plant PSI-ferredoxin supercomplex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane complex / photosystem I / ferredoxin / light harvesting / excitation transfer / PHOTOSYNTHESIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationphotosynthesis, light harvesting in photosystem I / photosynthesis, light harvesting / chloroplast thylakoid lumen / photosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / photosystem II / chloroplast stroma / chlorophyll binding ...photosynthesis, light harvesting in photosystem I / photosynthesis, light harvesting / chloroplast thylakoid lumen / photosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / photosystem II / chloroplast stroma / chlorophyll binding / chloroplast thylakoid membrane / photosynthesis / electron transport chain / 2 iron, 2 sulfur cluster binding / 4 iron, 4 sulfur cluster binding / electron transfer activity / oxidoreductase activity / protein domain specific binding / magnesium ion binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Pisum sativum (garden pea) Pisum sativum (garden pea) | |||||||||
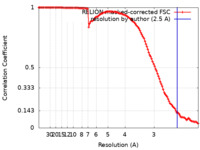
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Caspy I / Nelson N | |||||||||
| Funding support |  Israel, 2 items Israel, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2020 Journal: Nat Plants / Year: 2020Title: The structure of a triple complex of plant photosystem I with ferredoxin and plastocyanin. Authors: Ido Caspy / Anna Borovikova-Sheinker / Daniel Klaiman / Yoel Shkolnisky / Nathan Nelson /  Abstract: The ability of photosynthetic organisms to use sunlight as a sole source of energy is endowed by two large membrane complexes-photosystem I (PSI) and photosystem II (PSII). PSI and PSII are the ...The ability of photosynthetic organisms to use sunlight as a sole source of energy is endowed by two large membrane complexes-photosystem I (PSI) and photosystem II (PSII). PSI and PSII are the fundamental components of oxygenic photosynthesis, providing oxygen, food and an energy source for most living organisms on Earth. Currently, high-resolution crystal structures of these complexes from various organisms are available. The crystal structures of megadalton complexes have revealed excitation transfer and electron-transport pathways within the various complexes. PSI is defined as plastocyanin-ferredoxin oxidoreductase but a high-resolution structure of the entire triple supercomplex is not available. Here, using a new cryo-electron microscopy technique, we solve the structure of native plant PSI in complex with its electron donor plastocyanin and the electron acceptor ferredoxin. We reveal all of the contact sites and the modes of interaction between the interacting electron carriers and PSI. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10746.map.gz emd_10746.map.gz | 128.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10746-v30.xml emd-10746-v30.xml emd-10746.xml emd-10746.xml | 39.2 KB 39.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10746_fsc.xml emd_10746_fsc.xml | 11.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_10746.png emd_10746.png | 99.7 KB | ||
| Filedesc metadata |  emd-10746.cif.gz emd-10746.cif.gz | 9.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10746 http://ftp.pdbj.org/pub/emdb/structures/EMD-10746 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10746 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10746 | HTTPS FTP |
-Related structure data
| Related structure data |  6yacMC  6yezC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10746.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10746.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.085 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
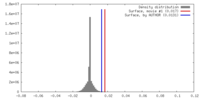
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Pea Photosystem I in complex with Ferredoxin
+Supramolecule #1: Pea Photosystem I in complex with Ferredoxin
+Macromolecule #1: Photosystem I P700 chlorophyll a apoprotein A1
+Macromolecule #2: Photosystem I P700 chlorophyll a apoprotein A2
+Macromolecule #3: Photosystem I iron-sulfur center
+Macromolecule #4: PsaD
+Macromolecule #5: PsaE
+Macromolecule #6: PsaF
+Macromolecule #7: PsaG
+Macromolecule #8: PsaH
+Macromolecule #9: Photosystem I reaction center subunit VIII
+Macromolecule #10: Photosystem I reaction center subunit IX
+Macromolecule #11: Photosystem I reaction center subunit X psaK
+Macromolecule #12: PsaL
+Macromolecule #13: Lhca1
+Macromolecule #14: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #15: Chlorophyll a-b binding protein 3, chloroplastic
+Macromolecule #16: Chlorophyll a-b binding protein P4, chloroplastic
+Macromolecule #17: Ferredoxin-1, chloroplastic
+Macromolecule #18: CHLOROPHYLL A ISOMER
+Macromolecule #19: CHLOROPHYLL A
+Macromolecule #20: PHYLLOQUINONE
+Macromolecule #21: IRON/SULFUR CLUSTER
+Macromolecule #22: BETA-CAROTENE
+Macromolecule #23: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #24: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #25: 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE
+Macromolecule #26: CALCIUM ION
+Macromolecule #27: DIGALACTOSYL DIACYL GLYCEROL (DGDG)
+Macromolecule #28: (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL
+Macromolecule #29: CHLOROPHYLL B
+Macromolecule #30: (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BE...
+Macromolecule #31: (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},...
+Macromolecule #32: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #33: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 3669 / Average electron dose: 1.075 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.9 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)